Help Instruction
- Double clicking on the substrate will redirect to the uniprot entry.
- Clicking in the block having the substrate will zoom into its phophosites
- Clicking the center of the circle will lead to zooming out.
- Clicking on the phosphosite will render the phosphosite in its 3D structure (if structure information/mapping is available).
- The color of every substrate has ben kept different in the Sunburst diagram for visual clarity.
- Clicking in the top right corner on the camara icon will download the image.
| Gene Name | Organism | P-Site | Sequence(+/-7) | Conservation | Disorder | Curator Assessment | Reliability | Evidence Class | Evidence Logic | PubMed | Phospho-ELM | PhosphoSite-Plus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MARK2 (Q8WUI4) | Homo sapiens | S155 | FPLRKTVSEPNLKLR | 0.972 | 0.6227 | TP | certain | experimental | 3 | 55 | 73 | 9 | 72 | 143 | | 16980613 | - |
|
| MARK1 (Q8WUI4) | Homo sapiens | S155 | FPLRKTVSEPNLKLR | 0.972 | 0.6227 | - | - | - | - | - | - | - |
| MARK3 (Q8WUI4) | Homo sapiens | S155 | FPLRKTVSEPNLKLR | 0.972 | 0.6227 | - | - | - | - | - | - | - |
| MARK4 (Q8WUI4) | Homo sapiens | S155 | FPLRKTVSEPNLKLR | 0.972 | 0.6227 | - | - | - | - | - | - | - |
| PRKD1 (Q8WUI4) | Homo sapiens | S155 | FPLRKTVSEPNLKLR | 0.972 | 0.6227 | - | - | - | - | - | Non high throughput (15623513, 18509061, 18617643) |
|
| CAMK1 (Q8WUI4) | Homo sapiens | S508 | SQARVLSSSETPART | 0.206 | 0.6531 | - | - | - | - | - | - | - |
| PRKD1 (Q8WUI4) | Homo sapiens | S358 | WPLSRTRSEPLPPSA | 0.984 | 0.8013 | - | - | - | - | - | - |
|
| PRKD1 (Q8WUI4) | Homo sapiens | S486 | RPLSRAQSSPAAPAS | 0.983 | 0.805 | - | - | - | - | - | - |
|
Help Instructions
- Hovering on the "*" in "Kinase Gene Name" column will display the text if there is a note by the annotator for the given site.
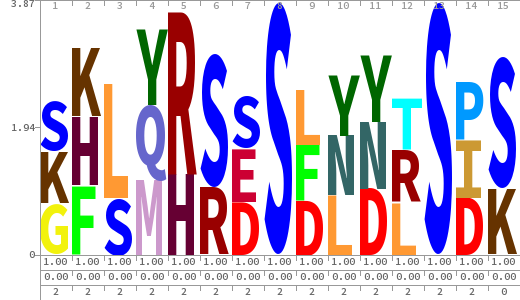
- The column having "Sequence" motif represents phosphosite and its neighbouring 7 residues (upstream and downstream).
- The score in "Conservation" column varies from 0 to 1; Higher the score more conserved is the site.
- The score in "Disorder" column varies from 0 to 1; Score higher than 0.4 suggests disorder.
- In the "Curator Assessment" column, TP refers to True Positive; FP refers to False Positive; and U refers to unknown if the annoatator cannot decide the status based on available information.
- Hovering on the number in the "Evidence Logic" column will show the name of method used to characterize/infer the phosphosite (for more details plase follow the link)