Help Instruction
- Double clicking on the substrate will redirect to the uniprot entry.
- Clicking in the block having the substrate will zoom into its phophosites
- Clicking the center of the circle will lead to zooming out.
- Clicking on the phosphosite will render the phosphosite in its 3D structure (if structure information/mapping is available).
- The color of every substrate has ben kept different in the Sunburst diagram for visual clarity.
- Clicking in the top right corner on the camara icon will download the image.
| Substrate | Organism | PSite | Sequence(+/-7) | Conservation | Disorder | Curator Assessment | Reliability | Evidence Class | Evidence Logic | PubMed | Phospho-ELM | PhosphoSite-Plus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RIN1 (Q13671) | Homo sapiens | S292 | QLLRRESSVGYRVPA | 0.737 | 0.5456 | TP | certain | experimental | 72 | 73 | 55 | 140 | | 21209314 | - |
|
| MARK2 * (Q7KZI7 ) | Homo sapiens | S400 | HKVQRSVSANPKQRR | 0.995 | 0.8279 | TP | likely | experimental | 140 | | 19011111 | - |
|
| SSH1 (Q8WYL5) | Homo sapiens | S937 | SNLTRSSSSDSIHSV | 0.839 | 0.6948 | TP | certain | experimental | 73 | 55 | 8 | | 19567672 | - | - |
| SSH1 (Q8WYL5) | Homo sapiens | S978 | SPLKRSHSLAKLGSL | 0.802 | 0.4369 | TP | certain | experimental | 73 | 55 | 8 | | 19567672 | - | - |
| PRKD2 (Q9BZL6) | Homo sapiens | S876 | QGLAERISVL_____ | 0.500 | 0.3053 | TP | certain | experimental | 73 | 55 | 9 | | 11062248, 12058027 | - |
|
| HDAC5 * (Q9UQL6 ) | Homo sapiens | S498 | RPLSRTQSSPLPQSP | 0.940 | 0.9126 | TP | likely | experimental | 73 | | 16584705 | - |
|
| PI4KB (Q9UBF8) | Homo sapiens | S294 | SNLKRTASNPKVENE | 0.332 | 0.7459 | TP | certain | experimental | 72 | 9 | 55 | 73 | 74 | | 16100512 | - | - |
| PKD2 (Q13563) | Homo sapiens | S801 | SSLPRPMSSRSFPRS | 0.451 | 0.6576 | TP | certain | experimental | 148 | 9 | 55 | 72 | 73 | | 20881056 | - | - |
| SET (Q01105-2) | Homo sapiens | S171 | KDLTKRSSQTQNKAS | 0.966 | 0.6089 | TP | certain | experimental | 148 | 72 | 73 | 9 | | 23251465 | - | - |
| CIB1 (Q99828-2) | Homo sapiens | S118 | ERICRVFSTSPAKDS | 0.788 | 0.1914 | TP | certain | experimental | 72 | 9 | 131 | 55 | 23 | | 23503467 | - | - |
| PI4KB (Q9UBF8-2) | Homo sapiens | S294 | SNLKRTASNPKVENE | 0.332 | 0.7547 | - | - | - | - | - | - |
|
| VASP (P50552) | Homo sapiens | S239 | GAKLRKVSKQEEASG | 0.577 | 0.892 | - | - | - | - | - | - |
|
| VASP (P50552) | Homo sapiens | S157 | EHIERRVSNAGGPPA | 0.401 | 0.9473 | - | - | - | - | - | - |
|
| CIB1 (Q99828) | Homo sapiens | S78 | ERICRVFSTSPAKDS | 0.873 | 0.1583 | - | - | - | - | - | - |
|
| VASP (P50552) | Homo sapiens | S322 | TTLPRMKSSSSVTTS | 0.364 | 0.8596 | - | - | - | - | - | - |
|
| ITGB4 (P16144-2) | Homo sapiens | T1736 | EFVSRTLTTSGTLST | 0.859 | 0.4864 | - | - | - | - | - | - |
|
| IFNAR1 (P17181) | Homo sapiens | S535 | SSQTSQDSGNYSNED | 0.965 | 0.6712 | - | - | - | - | - | - | - |
| RTKN (Q9BST9) | Homo sapiens | S448 | QALAKQGSLYHEMAI | 0.952 | 0.4979 | - | - | - | - | - | - | - |
Help Instructions
- Hovering on the "*" in "Substrate" column will display the text if there is a note by the annotator for the given site.
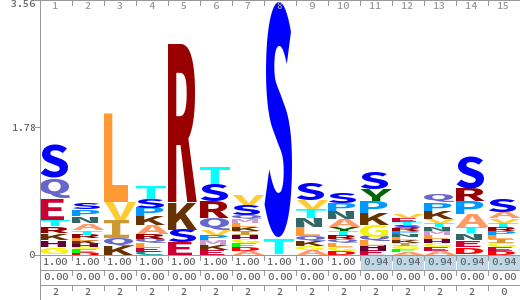
- The column having "Sequence" motif represents phosphosite and its neighbouring 7 residues (upstream and downstream).
- The score in "Conservation" column varies from 0 to 1; Higher the score more conserved is the site.
- The score in "Disorder" column varies from 0 to 1; Score higher than 0.4 suggests disorder.
- In the "Curator Assessment" column, TP refers to True Positive; FP refers to False Positive; and U refers to unknown if the annoatator cannot decide the status based on available information.
- Hovering on the number in the "Evidence Logic" column will show the name of method used to characterize/infer the phosphosite (for more details plase follow the link)