Help Instruction
- Double clicking on the substrate will redirect to the uniprot entry.
- Clicking in the block having the substrate will zoom into its phophosites
- Clicking the center of the circle will lead to zooming out.
- Clicking on the phosphosite will render the phosphosite in its 3D structure (if structure information/mapping is available).
- The color of every substrate has ben kept different in the Sunburst diagram for visual clarity.
- Clicking in the top right corner on the camara icon will download the image.
| Substrate | Organism | PSite | Sequence(+/-7) | Conservation | Disorder | Curator Assessment | Reliability | Evidence Class | Evidence Logic | PubMed | Phospho-ELM | PhosphoSite-Plus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP53 (P04637) | Homo sapiens | S392 | FKTEGPDSD______ | 0.808 | 0.7672 | TP | certain | not annotated | 74 | 8 | 72 | 73 | 55 | | 17108107 | - |
|
| TP53 (P04637) | Homo sapiens | S15 | PSVEPPLSQETFSDL | 0.830 | 0.708 | TP | certain | experimental | 74 | 8 | 72 | 73 | 55 | | 17108107 | - |
|
| MBP (P25274) | Oryctolagus cuniculus | T65 | KDHAARTTHYGSLPQ | - | 0.8198 | U | likely | experimental | 135 | 273 | 73 | | 12805220 | - |
|
| Prkaa1 * (P54645 ) | Rattus norvegicus | T183 | SDGEFLRTSCGSPNY | - | 0.2884 | TP | likely | experimental | 73 | 55 | | 14614828 | - |
|
| MELK (Q14680) | Homo sapiens | T167 | NKDYHLQTCCGSLAY | 0.843 | 0.1424 | TP | certain | experimental | 214 | 8 | | 14976552 | Non high throughput (14976552) |
|
| STK11 (Q15831) | Homo sapiens | T189 | LLLTTGGTLKISDLG | 0.486 | 0.2129 | TP | certain | experimental | 73 | 8 | | 11430832 | Non high throughput (11430832) |
|
| BRSK1 (Q8TDC3-2) | Homo sapiens | T205 | VGDSLLETSCGSPHY | 0.678 | 0.1702 | TP | certain | experimental | 214 | 8 | | 14976552 | Non high throughput (14976552) |
|
| PTEN (P60484) | Homo sapiens | S380 | EPDHYRYSDTTDSDP | 0.594 | 0.8966 | U | likely | experimental | 72 | 9 | 55 | | 15987703 | - | - |
| PTEN (P60484) | Homo sapiens | S385 | RYSDTTDSDPENEPF | 0.894 | 0.8279 | U | likely | experimental | 72 | 9 | 55 | | 15987703 | - | - |
| MARK3 (P27448) | Homo sapiens | S215 | KLDTFCGSPPYAAPE | 0.605 | 0.1643 | TP | certain | experimental | 9 | 55 | 30 | 133 | 135 | 72 | 73 | | 14976552 | - | - |
| PTEN (P60484) | Homo sapiens | T382 | DHYRYSDTTDSDPEN | 0.540 | 0.8493 | U | likely | experimental | 72 | 9 | 55 | | 15987703 | - | - |
| PRKAA2 (P54646) | Homo sapiens | T172 | SDGEFLRTSCGSPNY | 0.703 | 0.2715 | TP | certain | experimental | 72 | 55 | 73 | 23 | | 14985505 | - | - |
| PTEN (P60484) | Homo sapiens | T383 | HYRYSDTTDSDPENE | 0.261 | 0.8493 | U | likely | experimental | 72 | 9 | 55 | | 15987703 | - | - |
| SNRK (Q9NRH2) | Homo sapiens | T173 | QPGKKLTTSCGSLAY | 0.690 | 0.2817 | TP | certain | experimental | 8 | 135 | 30 | 72 | | 15733851 | - | - |
| STK11 (Q15831) | Homo sapiens | T185 | KPGNLLLTTGGTLKI | 0.389 | 0.2129 | TP | certain | experimental | 30 | 72 | 133 | 135 | | 12805220 | - | - |
| STK11 (Q15831) | Homo sapiens | T402 | TEAAQLSTKSRAEGR | 0.175 | 0.8198 | TP | certain | experimental | 30 | 72 | 133 | 135 | | 12805220 | - | - |
| STK11 (Q15831) | Homo sapiens | T336 | KDRWRSMTVVPYLED | 0.974 | 0.4541 | TP | certain | experimental | 72 | 55 | 30 | 133 | 135 | | 12805220 | - | - |
| STK11 (Q15831) | Homo sapiens | T363 | IEDDIIYTQDFTVPG | 0.391 | 0.346 | TP | certain | experimental | 72 | 55 | 30 | 133 | 135 | | 12805220 | - | - |
| BRSK1 (Q8TDC3) | Homo sapiens | T189 | VGDSLLETSCGSPHY | 0.685 | 0.1702 | TP | certain | experimental | 9 | 55 | 72 | 73 | | 14976552 | - | - |
| BRSK2 (Q8IWQ3) | Homo sapiens | T174 | VGDSLLETSCGSPHY | 0.628 | 0.1671 | TP | certain | experimental | 9 | 55 | 30 | 135 | 72 | 73 | | 14976552 | - | - |
| MARK1 (Q9P0L2) | Homo sapiens | T215 | TVGNKLDTFCGSPPY | 0.700 | 0.2503 | TP | certain | experimental | 9 | 55 | 72 | 73 | | 14976552 | - | - |
| MARK2 (Q7KZI7) | Homo sapiens | T208 | TFGNKLDTFCGSPPY | 0.633 | 0.1969 | TP | certain | experimental | 9 | 55 | 72 | 73 | | 14976552 | - | - |
| MARK3 (P27448) | Homo sapiens | T211 | TVGGKLDTFCGSPPY | 0.642 | 0.2399 | TP | certain | experimental | 9 | 55 | 30 | 133 | 135 | 72 | 73 | | 14976552 | - | - |
| MARK4 (Q96L34) | Homo sapiens | T214 | TLGSKLDTFCGSPPY | 0.759 | 0.1969 | TP | certain | experimental | 9 | 55 | 30 | 133 | 135 | 72 | 73 | | 14976552 | - | - |
| NUAK2 (Q9H093) | Homo sapiens | T208 | HQGKFLQTFCGSPLY | 0.754 | 0.087 | TP | certain | experimental | 9 | 55 | 30 | 133 | 135 | 72 | 73 | | 14976552 | - | - |
| NUAK1 (O60285) | Homo sapiens | T211 | QKDKFLQTFCGSPLY | 0.765 | 0.0967 | TP | certain | experimental | 9 | 55 | 72 | 73 | | 14976552 | - | - |
| SIK1 (P57059) | Homo sapiens | T182 | KSGEPLSTWCGSPPY | 0.694 | 0.2094 | TP | certain | experimental | 9 | 55 | 30 | 133 | 135 | 72 | 73 | | 14976552 | - | - |
| SIK2 (Q9H0K1) | Homo sapiens | T175 | KSGELLATWCGSPPY | 0.707 | 0.0749 | TP | certain | experimental | 9 | 55 | 72 | 73 | | 14976552 | - | - |
| STRADA (Q7RTN6) | Homo sapiens | T329 | GLSDSLTTSTPRPSN | 0.110 | 0.7982 | TP | certain | experimental | 23 | 72 | 73 | 30 | 133 | 135 | 9 | | 12805220 | - | - |
| STRADA (Q7RTN6) | Homo sapiens | T419 | SGIFGLVTNLEELEV | 0.145 | 0.3392 | TP | certain | experimental | 23 | 72 | 73 | 30 | 133 | 135 | 9 | | 12805220 | - | - |
| SMAD4 (Q13485) | Homo sapiens | T77 | KCVTIQRTLDGRLQV | 0.728 | 0.3566 | TP | certain | experimental | 72 | 135 | 62 | | 20974850 | - | - |
| PAK1 (Q13153) | Homo sapiens | T109 | QWARLLQTSNITKSE | 0.052 | 0.6806 | TP | certain | experimental | 72 | 73 | 9 | 30 | | 20400510 | - | - |
| SIK3 (J3KPC8) | Homo sapiens | T221 | TPGQLLKTWCGSPPY | 0.456 | 0.1137 | TP | certain | experimental | 9 | 55 | 72 | 73 | | 14976552 | - | - |
| Stk11 (Q9WTK7) | Mus musculus | T336 | KDRWRSMTVVPYLED | - | 0.433 | - | - | - | - | - | Non high throughput (11853558) | - |
| Stk11 (Q9WTK7) | Mus musculus | T366 | IEDGIIYTQDFTVPG | - | 0.2224 | - | - | - | - | - | Non high throughput (11853558) | - |
| SIK3 (Q9Y2K2) | Homo sapiens | T221 | TPGQLLKTWCGSPPY | 0.456 | 0.1137 | - | - | - | - | - | Non high throughput (14976552) | - |
| MARK3 (P27448-2) | Homo sapiens | T234 | TVGGKLDTFCGSPPY | 0.499 | 0.2258 | - | - | - | - | - | Non high throughput (14976552) | - |
| STK11 (Q15831) | Homo sapiens | S428 | SSKIRRLSACKQQ__ | 0.975 | 0.3774 | - | - | - | - | - | - |
|
| Sik1 (Q9R1U5) | Rattus norvegicus | T182 | KPGEPLSTWCGSPPY | - | 0.2609 | - | - | - | - | - | - |
|
| ETV4 (P43268) | Homo sapiens | S396 | AMNYDKLSRSLRYYY | 0.612 | 0.2258 | - | - | - | - | - | - | - |
| GSK3B (P49841) | Homo sapiens | S9 | SGRPRTTSFAESCKP | 0.500 | 0.5951 | - | - | - | - | - | - | - |
| PRKAA2 (P54646) | Homo sapiens | T485 | EQRSGSSTPQRSCSA | 0.154 | 0.5176 | - | - | - | - | - | - | - |
| SIK1B (A0A0B4J2F2) | Homo sapiens | T182 | KSGEPLSTWCGSPPY | - | 0.2094 | - | - | - | - | - | - | - |
| (Q9HBS3) | Homo sapiens | T336 | KDRWRSMTVVPYLED | - | 0.4507 | - | - | - | - | - | - | - |
| STK11 (A0A0S2Z4D1) | Homo sapiens | T189 | LLLTTGGTLKISDLG | - | 0.2129 | - | - | - | - | - | - | - |
| SNRK (A0A024R2Y6) | Homo sapiens | T173 | QPGKKLTTSCGSLAY | - | 0.2817 | - | - | - | - | - | - | - |
| MARK1 (B4DIB3) | Homo sapiens | T215 | TVGNKLDTFCGSPPY | - | 0.2503 | - | - | - | - | - | - | - |
Help Instructions
- Hovering on the "*" in "Substrate" column will display the text if there is a note by the annotator for the given site.
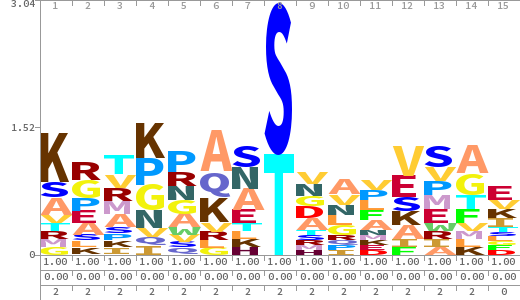
- The column having "Sequence" motif represents phosphosite and its neighbouring 7 residues (upstream and downstream).
- The score in "Conservation" column varies from 0 to 1; Higher the score more conserved is the site.
- The score in "Disorder" column varies from 0 to 1; Score higher than 0.4 suggests disorder.
- In the "Curator Assessment" column, TP refers to True Positive; FP refers to False Positive; and U refers to unknown if the annoatator cannot decide the status based on available information.
- Hovering on the number in the "Evidence Logic" column will show the name of method used to characterize/infer the phosphosite (for more details plase follow the link)