Help Instruction
- Double clicking on the substrate will redirect to the uniprot entry.
- Clicking in the block having the substrate will zoom into its phophosites
- Clicking the center of the circle will lead to zooming out.
- Clicking on the phosphosite will render the phosphosite in its 3D structure (if structure information/mapping is available).
- The color of every substrate has ben kept different in the Sunburst diagram for visual clarity.
- Clicking in the top right corner on the camara icon will download the image.
| Substrate | Organism | PSite | Sequence(+/-7) | Conservation | Disorder | Curator Assessment | Reliability | Evidence Class | Evidence Logic | PubMed | Phospho-ELM | PhosphoSite-Plus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPRY2 (O43597) | Homo sapiens | S112 | APLSRSISTVSSGSR | 0.354 | 0.7036 | TP | certain | experimental | 140 | 148 | 8 | | 19864419 | - |
|
| SPRY2 (O43597) | Homo sapiens | S121 | VSSGSRSSTRTSTSS | 0.854 | 0.7289 | TP | certain | experimental | 140 | 148 | 8 | | 19864419 | - |
|
| SFPQ (P23246) | Homo sapiens | S8 | MSRDRFRSRGGGGGG | 0.448 | 0.9913 | TP | certain | experimental | 72 | 107 | 30 | 9 | 55 | 148 | | 17965020 | - | - |
| SFPQ (P23246) | Homo sapiens | S283 | EGFKANLSLLRRPGE | 0.784 | 0.6755 | TP | certain | experimental | 72 | 107 | 30 | 9 | 148 | | 17965020 | - | - |
| PLEC (Q15149) | Homo sapiens | S4642 | RAGSRRGSFDATGSG | 0.689 | 0.5139 | TP | certain | experimental | 148 | 55 | | 23843618 | - | - |
| EIF4E * (P06730 ) | Homo sapiens | S209 | DTATKSGSTTKNRFV | 0.815 | 0.4507 | TP | certain | experimental | 72 | 137 | 133 | 147 | 73 | | 11154262 | - | - |
| HNRNPA1 (P09651) | Homo sapiens | S192 | EMASASSSQRGRSGS | 0.141 | 0.725 | - | - | - | - | - | - | - |
| HNRNPA1 (P09651) | Homo sapiens | S362 | GGYGGSSSSSSYGSG | 0.119 | 0.6269 | - | - | - | - | - | - | - |
| HNRNPA1 (P09651) | Homo sapiens | S363 | GYGGSSSSSSYGSGR | 0.046 | 0.5951 | - | - | - | - | - | - | - |
| ADRB2 (P07550) | Homo sapiens | S355 | KAYGNGYSSNGNTGE | 0.530 | 0.4582 | - | - | - | - | - | - | - |
| ADRB2 (P07550) | Homo sapiens | S356 | AYGNGYSSNGNTGEQ | 0.173 | 0.3774 | - | - | - | - | - | - | - |
| CHRM2 (P08172) | Homo sapiens | S286 | EKESSNDSTSVSAVA | 0.892 | 0.7982 | - | - | - | - | - | - | - |
| CHRM2 (P08172) | Homo sapiens | S288 | ESSNDSTSVSAVASN | 0.990 | 0.805 | - | - | - | - | - | - | - |
| CHRM2 (P08172) | Homo sapiens | S283 | QGEEKESSNDSTSVS | 0.516 | 0.6482 | - | - | - | - | - | - | - |
| CHRM2 (P08172) | Homo sapiens | S234 | NQDPVSPSLVQGRIV | 0.936 | 0.5176 | - | - | - | - | - | - | - |
| CHRM2 (P08172) | Homo sapiens | S290 | SNDSTSVSAVASNMR | 0.990 | 0.7982 | - | - | - | - | - | - | - |
| CHRM2 (P08172) | Homo sapiens | S232 | VANQDPVSPSLVQGR | 0.709 | 0.4901 | - | - | - | - | - | - | - |
| CHRM2 (P08172) | Homo sapiens | S294 | TSVSAVASNMRDDEI | 0.615 | 0.7459 | - | - | - | - | - | - | - |
| CHRM2 (P08172) | Homo sapiens | S250 | PNNNNMPSSDDGLEH | 0.816 | 0.6482 | - | - | - | - | - | - | - |
| CHRM2 (P08172) | Homo sapiens | S251 | NNNNMPSSDDGLEHN | 0.679 | 0.7547 | - | - | - | - | - | - | - |
| CHRM2 (P08172) | Homo sapiens | S309 | TQDENTVSTSLGHSK | 0.402 | 0.7916 | - | - | - | - | - | - | - |
| CHRM2 (P08172) | Homo sapiens | S282 | VQGEEKESSNDSTSV | 0.604 | 0.6806 | - | - | - | - | - | - | - |
| CHRM2 (P08172) | Homo sapiens | S311 | DENTVSTSLGHSKDE | 0.340 | 0.7672 | - | - | - | - | - | - | - |
| MC4R (P32245) | Homo sapiens | S329 | LGGLCDLSSRY____ | 0.124 | 0.0042 | - | - | - | - | - | - | - |
| MC4R (P32245) | Homo sapiens | S330 | GGLCDLSSRY_____ | 0.093 | 0.0059 | - | - | - | - | - | - | - |
| CCR5 (P51681) | Homo sapiens | S336 | QEAPERASSVYTRST | 0.802 | 0.2752 | - | - | - | - | - | - | - |
| CCR5 (P51681) | Homo sapiens | S337 | EAPERASSVYTRSTG | 0.974 | 0.3807 | - | - | - | - | - | - | - |
| CCR5 (P51681) | Homo sapiens | S349 | STGEQEISVGL____ | 0.726 | 0.2034 | - | - | - | - | - | - | - |
| CCR5 (P51681) | Homo sapiens | S342 | ASSVYTRSTGEQEIS | 0.835 | 0.268 | - | - | - | - | - | - | - |
| ADRB2 (P07550) | Homo sapiens | S411 | RNCSTNDSLL_____ | 0.560 | 0.3872 | - | - | - | - | - | - | - |
| ADRB2 (P07550) | Homo sapiens | S364 | NGNTGEQSGYHVEQE | 0.174 | 0.5854 | - | - | - | - | - | - | - |
| ADRB2 (P07550) | Homo sapiens | S401 | VPSDNIDSQGRNCST | 0.543 | 0.6043 | - | - | - | - | - | - | - |
| ADRB2 (P07550) | Homo sapiens | S407 | DSQGRNCSTNDSLL_ | 0.721 | 0.4186 | - | - | - | - | - | - | - |
| ADRA1B (P35368) | Homo sapiens | S410 | RKDSLDDSGSCLSGS | 0.859 | 0.562 | - | - | - | - | - | - | - |
| ADRA1B (P35368) | Homo sapiens | S412 | DSLDDSGSCLSGSQR | 0.573 | 0.5055 | - | - | - | - | - | - | - |
| ADRA1B (P35368) | Homo sapiens | S406 | RSQSRKDSLDDSGSC | 0.847 | 0.4541 | - | - | - | - | - | - | - |
| CHRM2 (P08172) | Homo sapiens | T302 | NMRDDEITQDENTVS | 0.144 | 0.6948 | - | - | - | - | - | - | - |
| CHRM2 (P08172) | Homo sapiens | T287 | KESSNDSTSVSAVAS | 0.994 | 0.7951 | - | - | - | - | - | - | - |
| CHRM2 (P08172) | Homo sapiens | T271 | KAPRDPVTENCVQGE | 0.152 | 0.7951 | - | - | - | - | - | - | - |
| CHRM2 (P08172) | Homo sapiens | T307 | EITQDENTVSTSLGH | 0.173 | 0.7916 | - | - | - | - | - | - | - |
| CHRM2 (P08172) | Homo sapiens | T310 | QDENTVSTSLGHSKD | 0.374 | 0.7843 | - | - | - | - | - | - | - |
| MC4R (P32245) | Homo sapiens | T312 | RSQELRKTFKEIICC | 0.809 | 0.0058 | - | - | - | - | - | - | - |
| ADRB2 (P07550) | Homo sapiens | T384 | LCEDLPGTEDFVGHQ | 0.302 | 0.2258 | - | - | - | - | - | - | - |
| ADRB2 (P07550) | Homo sapiens | T408 | SQGRNCSTNDSLL__ | 0.612 | 0.3807 | - | - | - | - | - | - | - |
Help Instructions
- Hovering on the "*" in "Substrate" column will display the text if there is a note by the annotator for the given site.
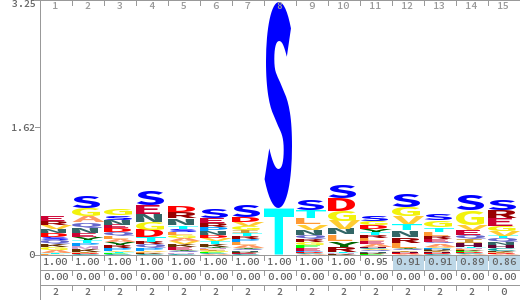
- The column having "Sequence" motif represents phosphosite and its neighbouring 7 residues (upstream and downstream).
- The score in "Conservation" column varies from 0 to 1; Higher the score more conserved is the site.
- The score in "Disorder" column varies from 0 to 1; Score higher than 0.4 suggests disorder.
- In the "Curator Assessment" column, TP refers to True Positive; FP refers to False Positive; and U refers to unknown if the annoatator cannot decide the status based on available information.
- Hovering on the number in the "Evidence Logic" column will show the name of method used to characterize/infer the phosphosite (for more details plase follow the link)