Help Instruction
- Double clicking on the substrate will redirect to the uniprot entry.
- Clicking in the block having the substrate will zoom into its phophosites
- Clicking the center of the circle will lead to zooming out.
- Clicking on the phosphosite will render the phosphosite in its 3D structure (if structure information/mapping is available).
- The color of every substrate has ben kept different in the Sunburst diagram for visual clarity.
- Clicking in the top right corner on the camara icon will download the image.
| Substrate | Organism | PSite | Sequence(+/-7) | Conservation | Disorder | Curator Assessment | Reliability | Evidence Class | Evidence Logic | PubMed | Phospho-ELM | PhosphoSite-Plus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RELA (Q04206) | Homo sapiens | S276 | SMQLRRPSDRELSEP | 0.767 | 0.5296 | - | - | - | - | - | Non high throughput (12628924) |
|
| HIST1H2AG (P0C0S8) | Homo sapiens | S2 | ______MSGRGKQGG | 0.718 | 0.7595 | - | - | - | - | - | Non high throughput (15010469) |
|
| RPS6KA5 (O75582) | Homo sapiens | S212 | DETERAYSFCGTIEY | 0.818 | 0.1137 | - | - | - | - | - | Non high throughput (15568999) |
|
| RPS6KA5 (O75582) | Homo sapiens | S376 | EKLFQGYSFVAPSIL | 0.904 | 0.1162 | - | - | - | - | - | Non high throughput (15568999) |
|
| RPS6KA5 (O75582) | Homo sapiens | S381 | GYSFVAPSILFKRNA | 0.850 | 0.0327 | - | - | - | - | - | Non high throughput (15568999) |
|
| RPS6KA5 (O75582) | Homo sapiens | S750 | RRKMKKTSTSTETRS | 0.571 | 0.7289 | - | - | - | - | - | Non high throughput (15568999) |
|
| RPS6KA5 (O75582) | Homo sapiens | S752 | KMKKTSTSTETRSSS | 0.271 | 0.8235 | - | - | - | - | - | Non high throughput (15568999) |
|
| RPS6KA5 (O75582) | Homo sapiens | S758 | TSTETRSSSSESSHS | 0.482 | 0.8198 | - | - | - | - | - | Non high throughput (15568999) |
|
| Rela (Q04207) | Mus musculus | S276 | SMQLRRPSDRELSEP | - | 0.5254 | - | - | - | - | - | Non high throughput (16135789) |
|
| HMGN1 (P05114) | Homo sapiens | S7 | _MPKRKVSSAEGAAK | 0.202 | 0.9081 | - | - | - | - | - | - | - |
| TH (P07101-3) | Homo sapiens | S40 | RFIGRRQSLIEDARK | 0.090 | 0.5296 | - | - | - | - | - | - |
|
| SRF (P11831) | Homo sapiens | S103 | RGLKRSLSEMEIGMV | 0.283 | 0.494 | - | - | - | - | - | - |
|
| Nr4a1 (P12813) | Mus musculus | S354 | GRRGRLPSKPKQPPD | - | 0.7755 | - | - | - | - | - | - |
|
| Creb1 (P15337) | Rattus norvegicus | S133 | EILSRRPSYRKILND | - | 0.4979 | - | - | - | - | - | - |
|
| CREB1 (P16220) | Homo sapiens | S133 | EILSRRPSYRKILND | - | 0.4979 | - | - | - | - | - | - |
|
| ATF1 (P18846) | Homo sapiens | S63 | GILARRPSYRKILKD | 0.920 | 0.4864 | - | - | - | - | - | - |
|
| ELK1 (P19419) | Homo sapiens | S383 | IHFWSTLSPIAPRSP | 0.787 | 0.4801 | - | - | - | - | - | - |
|
| PLA2G4A (P47712) | Homo sapiens | S727 | RQNPSRCSVSLSNVE | 0.781 | 0.433 | - | - | - | - | - | - | - |
| HIST1H3A (P68431) | Homo sapiens | S11 | TKQTARKSTGGKAPR | 0.711 | 0.7505 | - | - | - | - | - | - |
|
| HIST1H3A (P68431) | Homo sapiens | S29 | ATKAARKSAPATGGV | 0.614 | 0.6375 | - | - | - | - | - | - |
|
| Hist1h3a (P68433) | Mus musculus | S11 | TKQTARKSTGGKAPR | - | 0.7505 | - | - | - | - | - | - |
|
| Hist1h3a (P68433) | Mus musculus | S29 | ATKAARKSAPATGGV | - | 0.6375 | - | - | - | - | - | - |
|
| H3F3A (P84243) | Homo sapiens | S11 | TKQTARKSTGGKAPR | 0.568 | 0.7629 | - | - | - | - | - | - | - |
| H3F3A (P84243) | Homo sapiens | S29 | ATKAARKSAPSTGGV | 0.558 | 0.6851 | - | - | - | - | - | - | - |
| Eif4ebp1 (Q60876) | Mus musculus | S64 | FLMECRNSPVAKTPP | - | 0.6712 | - | - | - | - | - | - |
|
| Bad (Q61337) | Mus musculus | S155 | GRELRRMSDEFEGSF | - | 0.5456 | - | - | - | - | - | - |
|
| Eif4ebp1 (Q60876) | Mus musculus | T36 | PPGDYSTTPGGTLFS | - | 0.7843 | - | - | - | - | - | - |
|
| WNK1 (Q9H4A3) | Homo sapiens | T60 | EYRRRRHTMDKDSRG | 0.599 | 0.8872 | - | - | - | - | - | - |
|
| Hist1h2ab (C0HKE1) | Mus musculus | S2 | ______MSGRGKQGG | - | 0.7672 | - | - | - | - | - | - |
|
| RARA (P10276) | Homo sapiens | S369 | YVRKRRPSRPHMFPK | 0.240 | 0.3704 | - | - | - | - | - | - |
|
| ATXN1 (P54253) | Homo sapiens | S775 | ATRKRRWSAPESRKL | 0.882 | 0.7289 | - | - | - | - | - | - |
|
| Trim28 (Q62318) | Mus musculus | S473 | SGMKRSRSGEGEVSG | - | 0.708 | - | - | - | - | - | - |
|
Help Instructions
- Hovering on the "*" in "Substrate" column will display the text if there is a note by the annotator for the given site.
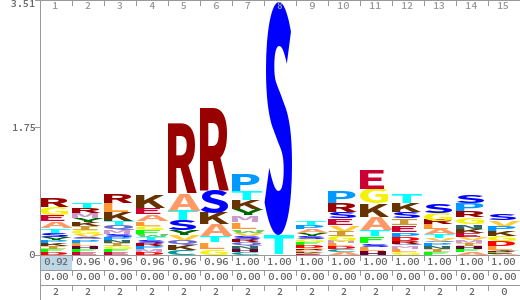
- The column having "Sequence" motif represents phosphosite and its neighbouring 7 residues (upstream and downstream).
- The score in "Conservation" column varies from 0 to 1; Higher the score more conserved is the site.
- The score in "Disorder" column varies from 0 to 1; Score higher than 0.4 suggests disorder.
- In the "Curator Assessment" column, TP refers to True Positive; FP refers to False Positive; and U refers to unknown if the annoatator cannot decide the status based on available information.
- Hovering on the number in the "Evidence Logic" column will show the name of method used to characterize/infer the phosphosite (for more details plase follow the link)