Help Instruction
- Double clicking on the substrate will redirect to the uniprot entry.
- Clicking in the block having the substrate will zoom into its phophosites
- Clicking the center of the circle will lead to zooming out.
- Clicking on the phosphosite will render the phosphosite in its 3D structure (if structure information/mapping is available).
- The color of every substrate has ben kept different in the Sunburst diagram for visual clarity.
- Clicking in the top right corner on the camara icon will download the image.
| Substrate | Organism | PSite | Sequence(+/-7) | Conservation | Disorder | Curator Assessment | Reliability | Evidence Class | Evidence Logic | PubMed | Phospho-ELM | PhosphoSite-Plus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tbc1d4 (Q8BYJ6) | Mus musculus | S711 | PSLHTSFSAPSFTAP | - | 0.3392 | TP | certain | experimental | 214 | 72 | 73 | 55 | 8 | | 19923418 | - |
|
| IKBKB (O14920) | Homo sapiens | S177 | AKELDQGSLCTSFVG | 0.636 | 0.087 | TP | certain | experimental | 73 | 62 | | 21673972 | - |
|
| IKBKB (O14920) | Homo sapiens | S181 | DQGSLCTSFVGTLQY | 0.873 | 0.0985 | TP | certain | experimental | 73 | 62 | | 21673972 | - |
|
| HDAC5 (Q9UQL6) | Homo sapiens | S259 | FPLRKTASEPNLKVR | 0.835 | 0.6712 | TP | certain | experimental | 73 | 8 | 23 | 55 | 72 | | 18184930 | - |
|
| HDAC5 (Q9UQL6) | Homo sapiens | S498 | RPLSRTQSSPLPQSP | 0.940 | 0.9126 | TP | certain | experimental | 73 | 8 | 23 | 55 | 72 | | 18184930 | - |
|
| FOXO3 * (O43524 ) | Homo sapiens | S399 | DNITLPPSQPSPTGG | 0.301 | 0.7672 | U | likely | experimental | 72 | 9 | | 17711846 | - | - |
| FOXO3 * (O43524 ) | Homo sapiens | S413 | GLMQRSSSFPYTTKG | 0.437 | 0.6227 | TP | certain | experimental | 72 | 9 | 31 | 55 | 73 | | 17711846 | - | - |
| FOXO3 * (O43524 ) | Homo sapiens | S555 | RALSNSVSNMGLSES | 0.204 | 0.5419 | U | likely | experimental | 72 | 9 | 31 | | 17711846 | - | - |
| FOXO3 * (O43524 ) | Homo sapiens | S588 | QTLSDSLSGSSLYST | 0.279 | 0.494 | U | likely | experimental | 72 | 9 | 55 | 73 | | 17711846 | - | - |
| FOXO3 * (O43524 ) | Homo sapiens | S626 | SLECDMESIIRSELM | 0.672 | 0.3529 | U | likely | experimental | 72 | 9 | 31 | 73 | | 17711846 | - | - |
| Vasp (P70460) | Mus musculus | S317 | TTLPRMKSSSSVTTS | - | 0.8623 | TP | certain | experimental | 72 | 9 | | 21945940 | - | - |
| PLD1 (Q13393) | Homo sapiens | S505 | GSVKRVTSGPSLGSL | 0.108 | 0.4901 | TP | certain | experimental | 23 | 72 | 20 | 8 | 73 | | 20231899 | - | - |
| FOXO3 (O43524) | Homo sapiens | T179 | SSPDKRLTLSQIYEW | 0.645 | 0.3807 | TP | likely | experimental | 72 | 9 | | 17711846 | - | - |
| HAS2 (Q92819) | Homo sapiens | T110 | LQSVKRLTYPGIKVV | 0.270 | 0.0281 | TP | certain | experimental | 55 | 9 | | 21228273 | - | - |
| SIRT1 (Q96EB6) | Homo sapiens | T344 | GKLLRNYTQNIDTLE | 0.231 | 0.2645 | TP | certain | experimental | 72 | 20 | 9 | 73 | | 22728651 | - | - |
| TP73 (O15350-1) | Homo sapiens | S426 | GGMNKLPSVNQLVGQ | 0.696 | 0.7036 | TP | certain | experimental | 72 | 9 | 73 | 23 | 55 | | 24874608 | - | - |
| Tp53 (P02340) | Mus musculus | S18 | ISLELPLSQETFSGL | - | 0.2988 | - | - | - | - | - | - |
|
| Ppargc1a (O70343) | Mus musculus | S538 | SLFDVSPSCSSFNSP | - | 0.3087 | - | - | - | - | - | - |
|
| Kcnb1 (P15387) | Rattus norvegicus | S444 | ERAKRNGSIVSMNMK | - | 0.3631 | - | - | - | - | - | - |
|
| Kcnb1 (P15387) | Rattus norvegicus | S541 | SKMAKTQSQPILNTK | - | 0.6227 | - | - | - | - | - | - |
|
| ACACA (Q13085) | Homo sapiens | S80 | LHIRSSMSGLHLVKQ | 0.956 | 0.2849 | - | - | - | - | - | - |
|
| Ppargc1a (O70343) | Mus musculus | T177 | NHTHRIRTNPAIVKT | - | 0.805 | - | - | - | - | - | - |
|
| PARP1 (P09874) | Homo sapiens | S177 | LGFRPEYSASQLKGF | 0.348 | 0.3087 | - | - | - | - | - | - |
|
| EEF2K (O00418) | Homo sapiens | S398 | DSLPSSPSSATPHSQ | 0.546 | 0.7163 | - | - | - | - | - | - |
|
| DUSP1 (P28562) | Homo sapiens | S334 | AVLDRGTSTTTVFNF | 0.129 | 0.2292 | - | - | - | - | - | - |
|
| Parp1 (P11103) | Mus musculus | S177 | LGFRPEYSASQLKGF | - | 0.3053 | - | - | - | - | - | - |
|
| GLI1 (P08151) | Homo sapiens | S408 | GPLPRAPSISTVEPK | 0.213 | 0.8792 | - | - | - | - | - | - |
|
| GLI1 (P08151) | Homo sapiens | S102 | LQTVIRTSPSSLVAF | 0.905 | 0.3704 | - | - | - | - | - | - |
|
| TFAP2A (P05549) | Homo sapiens | S219 | CSVPGRLSLLSSTSK | 0.841 | 0.1702 | - | - | - | - | - | - |
|
| Prkaa2 (Q8BRK8) | Mus musculus | T172 | SDGEFLRTSCGSPNY | - | 0.2752 | - | - | - | - | - | - |
|
| GLI1 (P08151) | Homo sapiens | T1074 | QRGSSGHTPPPSGPP | 0.304 | 0.9725 | - | - | - | - | - | - |
|
| PPARGC1A (Q9UBK2-3) | Homo sapiens | S527 | EDESDKLSYPWDGTQ | 0.870 | 0.5533 | - | - | - | - | - | - | - |
| EEF2K (O00418) | Homo sapiens | S366 | SPQVRTLSGSRPPLL | 0.742 | 0.6227 | - | - | - | - | - | - | - |
| EEF2K (O00418) | Homo sapiens | S78 | SSGSPANSFHFKEAW | 0.374 | 0.3667 | - | - | - | - | - | - | - |
| PFKFB2 (O60825) | Homo sapiens | S466 | PVRMRRNSFTPLSSS | 0.089 | 0.5807 | - | - | - | - | - | - | - |
| ULK1 (O75385) | Homo sapiens | S317 | SHLASPPSLGEMQQL | 0.841 | 0.7982 | - | - | - | - | - | - | - |
| ULK1 (O75385) | Homo sapiens | S556 | GLGCRLHSAPNLSDL | 0.945 | 0.5665 | - | - | - | - | - | - | - |
| ULK1 (O75385) | Homo sapiens | S638 | FDFPKTPSSQNLLAL | 0.789 | 0.4864 | - | - | - | - | - | - | - |
| NOS3 (P29474) | Homo sapiens | S1177 | TSRIRTQSFSLQERQ | 0.140 | 0.433 | - | - | - | - | - | - | - |
| NOS3 (P29474) | Homo sapiens | S633 | WRRKRKESSNTDSAG | 0.163 | 0.4725 | - | - | - | - | - | - | - |
| CTNNB1 (P35222) | Homo sapiens | S552 | QDTQRRTSMGGTQQQ | 0.142 | 0.6661 | - | - | - | - | - | - | - |
| ACACA (Q13085) | Homo sapiens | S78 | LALHIRSSMSGLHLV | 0.987 | 0.2951 | - | - | - | - | - | - | - |
| ACACA (Q13085-2) | Homo sapiens | S22 | MLQRSSMSGLHLVKQ | 0.000 | 0.2715 | - | - | - | - | - | - | - |
| ACACA (Q13085-4) | Homo sapiens | S117 | LHIRSSMSGLHLVKQ | 0.302 | 0.2541 | - | - | - | - | - | - | - |
| CRTC2 (Q53ET0) | Homo sapiens | S171 | SALNRTSSDSALHTS | 0.886 | 0.6531 | - | - | - | - | - | - | - |
| TBC1D1 (Q86TI0) | Homo sapiens | S237 | RPMRKSFSQPGLRSL | 0.733 | 0.422 | - | - | - | - | - | - | - |
| TP53 (P04637) | Homo sapiens | S15 | PSVEPPLSQETFSDL | 0.830 | 0.708 | - | - | - | - | - | - | - |
| KCNB1 (Q14721) | Homo sapiens | S444 | ERAKRNGSIVSMNMK | 0.881 | 0.3774 | - | - | - | - | - | - | - |
| KCNB1 (Q14721) | Homo sapiens | S541 | NKMAKTQSQPILNTK | 0.974 | 0.6755 | - | - | - | - | - | - | - |
| VASP (A0A024R0V4) | Homo sapiens | S322 | TTLPRMKSSSSVTTS | - | 0.8596 | - | - | - | - | - | - | - |
| TBC1D4 (A0A024R637) | Homo sapiens | S704 | PSLHTSFSAPSFTAP | - | 0.4507 | - | - | - | - | - | - | - |
| ACACB (O00763) | Homo sapiens | S222 | PTMRPSMSGLHLVKR | 0.966 | 0.6043 | - | - | - | - | - | - | - |
| PARP1 (A0A024R3T8) | Homo sapiens | S177 | LGFRPEYSASQLKGF | - | 0.3087 | - | - | - | - | - | - | - |
| FYN (P06241) | Homo sapiens | T12 | QCKDKEATKLTEERD | 0.189 | 0.4476 | - | - | - | - | - | - | - |
| PPARGC1A (Q9UBK2) | Homo sapiens | T166 | NECSGLSTQNHANHN | 0.120 | 0.6136 | - | - | - | - | - | - | - |
| (B2RA25) | Homo sapiens | T172 | SDGEFLRTSCGSPNY | - | 0.2715 | - | - | - | - | - | - | - |
Help Instructions
- Hovering on the "*" in "Substrate" column will display the text if there is a note by the annotator for the given site.
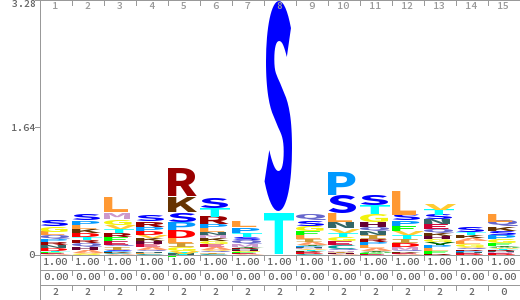
- The column having "Sequence" motif represents phosphosite and its neighbouring 7 residues (upstream and downstream).
- The score in "Conservation" column varies from 0 to 1; Higher the score more conserved is the site.
- The score in "Disorder" column varies from 0 to 1; Score higher than 0.4 suggests disorder.
- In the "Curator Assessment" column, TP refers to True Positive; FP refers to False Positive; and U refers to unknown if the annoatator cannot decide the status based on available information.
- Hovering on the number in the "Evidence Logic" column will show the name of method used to characterize/infer the phosphosite (for more details plase follow the link)