Help Instruction
- Double clicking on the substrate will redirect to the uniprot entry.
- Clicking in the block having the substrate will zoom into its phophosites
- Clicking the center of the circle will lead to zooming out.
- Clicking on the phosphosite will render the phosphosite in its 3D structure (if structure information/mapping is available).
- The color of every substrate has ben kept different in the Sunburst diagram for visual clarity.
- Clicking in the top right corner on the camara icon will download the image.
| Substrate | Organism | PSite | Sequence(+/-7) | Conservation | Disorder | Curator Assessment | Reliability | Evidence Class | Evidence Logic | PubMed | Phospho-ELM | PhosphoSite-Plus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bad (Q61337) | Mus musculus | S170 | KGLPRPKSAGTATQM | - | 0.8235 | TP | certain | experimental | 72 | 55 | 9 | 73 | | 22103330 | - | - |
| STAT3 (P40763) | Homo sapiens | S727 | NTIDLPMSPRTLDSL | 0.410 | 0.3599 | TP | certain | experimental | 72 | 73 | 23 | 55 | 148 | | 18483256 | - | - |
| RELA (Q04206) | Homo sapiens | S536 | SGDEDFSSIADMDFS | 0.319 | 0.2783 | TP | certain | experimental | 148 | 72 | 9 | 55 | 16 | 31 | | 24248603 | - | - |
| RARA (P10276) | Homo sapiens | T209 | LCQLGKYTTNNSSEQ | 0.418 | 0.247 | TP | certain | experimental | 23 | 40 | 72 | 73 | 66 | | 17431504 | - | - |
| RARA (P10276) | Homo sapiens | T210 | CQLGKYTTNNSSEQR | 0.261 | 0.268 | TP | certain | experimental | 23 | 40 | 72 | 73 | 66 | | 17431504 | - | - |
| CAMK2G (Q13555) | Homo sapiens | S280 | CQRSTVASMMHRQET | 0.616 | 0.3392 | - | - | - | - | - | - |
|
| CAMK2G (Q13555) | Homo sapiens | S319 | RNFSAAKSLLNKKSD | 0.036 | 0.4507 | - | - | - | - | - | - |
|
| DAB2 (P98082) | Homo sapiens | S723 | KPAPRQVSLPVTKST | 0.019 | 0.8655 | - | - | - | - | - | - |
|
| OCLN (Q16625) | Homo sapiens | S471 | LDDYREESEEYMAAA | 0.691 | 0.5758 | - | - | - | - | - | - |
|
| RRAD (P55042) | Homo sapiens | S273 | AGTRRRESLGKKAKR | 0.915 | 0.494 | - | - | - | - | - | - | - |
| BAD (Q92934) | Homo sapiens | S134 | KGLPRPKSAGTATQM | 0.882 | 0.8279 | - | - | - | - | - | - | - |
| (B3KVJ0) | Homo sapiens | S1051 | NVASRMESTGVMGNI | - | 0.3321 | - | - | - | - | - | - | - |
| (A9CB80) | Homo sapiens | S1070 | DSFLQRYSSDPTGAL | - | 0.5807 | - | - | - | - | - | - | - |
| (A9CB80) | Homo sapiens | S1071 | SFLQRYSSDPTGALT | - | 0.6712 | - | - | - | - | - | - | - |
| TH (P07101) | Homo sapiens | S19 | KGFRRAVSELDAKQA | 0.455 | 0.5665 | - | - | - | - | - | - | - |
| (B2R8K4) | Homo sapiens | S63 | GILARRPSYRKILRD | - | 0.5017 | - | - | - | - | - | - | - |
| RYR1 (P21817) | Homo sapiens | S2843 | KKKTRKISQSAQTYD | 0.703 | 0.6851 | - | - | - | - | - | - | - |
| STAT1 (P42224) | Homo sapiens | S727 | TDNLLPMSPEEFDEV | 0.902 | 0.4979 | - | - | - | - | - | - | - |
| GRIA1 (P42261) | Homo sapiens | S849 | FCLIPQQSINEAIRT | 0.238 | 0.1852 | - | - | - | - | - | - | - |
| PEA15 (Q15121) | Homo sapiens | S116 | KDIIRQPSEEEIIKL | 0.538 | 0.4017 | - | - | - | - | - | - | - |
| CREB1 (P16220) | Homo sapiens | S133 | EILSRRPSYRKILND | - | 0.4979 | - | - | - | - | - | - | - |
| CREB1 (P16220) | Homo sapiens | S142 | RKILNDLSSDAPGVP | - | 0.5707 | - | - | - | - | - | - | - |
| ATP2A2 (P16615) | Homo sapiens | S38 | KLKERWGSNELPAEE | 0.070 | 0.4149 | - | - | - | - | - | - | - |
| CEBPB (P17676) | Homo sapiens | S325 | EQLSRELSTLRNLFK | 0.002 | 0.5296 | - | - | - | - | - | - | - |
| FLNA (P21333) | Homo sapiens | S2523 | VTGPRLVSNHSLHET | 0.139 | 0.5055 | - | - | - | - | - | - | - |
| CFL2 (Q9Y281) | Homo sapiens | S24 | DMKVRKSSTQEEIKK | 0.207 | 0.3774 | - | - | - | - | - | - | - |
| DLG1 (Q12959) | Homo sapiens | S232 | ITLERGNSGLGFSIA | 0.126 | 0.3529 | - | - | - | - | - | - | - |
| ADCY3 (O60266) | Homo sapiens | S1076 | NVASRMESTGVMGNI | 0.605 | 0.3321 | - | - | - | - | - | - | - |
| KRT18 (P05783) | Homo sapiens | S53 | ISVSRSTSFRGGMGS | 0.418 | 0.4476 | - | - | - | - | - | - | - |
| QDPR (P09417) | Homo sapiens | S223 | ITGKNRPSSGSLIQV | 0.149 | 0.4087 | - | - | - | - | - | - | - |
| GFAP (P14136) | Homo sapiens | S38 | LGPGTRLSLARMPPP | 0.368 | 0.6851 | - | - | - | - | - | - | - |
| GFAP (P14136) | Homo sapiens | S13 | ITSAARRSYVSSGEM | 0.795 | 0.4292 | - | - | - | - | - | - | - |
| GFAP (P14136) | Homo sapiens | S17 | ARRSYVSSGEMMVGG | 0.894 | 0.4441 | - | - | - | - | - | - | - |
| HSF1 (Q00613) | Homo sapiens | S230 | PKYSRQFSLEHVHGS | 0.584 | 0.5055 | - | - | - | - | - | - | - |
| PLCB3 (Q01970) | Homo sapiens | S537 | PSLEPQKSLGDEGLN | 0.766 | 0.8623 | - | - | - | - | - | - | - |
| KCND2 (Q9NZV8) | Homo sapiens | S438 | ARIRAAKSGSANAYM | 0.059 | 0.346 | - | - | - | - | - | - | - |
| KCND2 (Q9NZV8) | Homo sapiens | S459 | LLSNQLQSSEDEQAF | 0.097 | 0.4369 | - | - | - | - | - | - | - |
| TH (P07101) | Homo sapiens | S44 | PGPSLTGSPWPGTAA | 0.066 | 0.8198 | - | - | - | - | - | - | - |
| SYN1 (P17600) | Homo sapiens | S605 | AGPTRQASQAGPVPR | 0.334 | 0.9905 | - | - | - | - | - | - | - |
| SYN1 (P17600) | Homo sapiens | S568 | PQATRQTSVSGPAPP | 0.924 | 0.9898 | - | - | - | - | - | - | - |
| ACACA (Q13085) | Homo sapiens | S25 | RFIIGSVSEDNSEDE | 0.000 | 0.4901 | - | - | - | - | - | - | - |
| GRIA1 (P42261) | Homo sapiens | S645 | LTVERMVSPIESAED | 0.029 | 0.1275 | - | - | - | - | - | - | - |
| VIM (P08670) | Homo sapiens | S39 | TTSTRTYSLGSALRP | 0.934 | 0.4652 | - | - | - | - | - | - | - |
| VIM (P08670) | Homo sapiens | S83 | GVRLLQDSVDFSLAD | 0.105 | 0.2849 | - | - | - | - | - | - | - |
| RYR2 (Q92736) | Homo sapiens | S2808 | YNRTRRISQTSQVSV | 0.753 | 0.346 | - | - | - | - | - | - | - |
| PPP3CA (A0A0S2Z4C6) | Homo sapiens | S411 | GKMARVFSVLREESE | - | 0.2258 | - | - | - | - | - | - | - |
| GRIN2B (Q13224) | Homo sapiens | S1303 | NKLRRQHSYDTFVDL | 0.872 | 0.5758 | - | - | - | - | - | - | - |
| GRIN2B (Q13224) | Homo sapiens | S383 | VGKWKDKSLQMKYYV | 0.724 | 0.1007 | - | - | - | - | - | - | - |
| OPRM1 (P35372) | Homo sapiens | S268 | LKSVRMLSGSKEKDR | 0.385 | 0.0909 | - | - | - | - | - | - | - |
| OPRM1 (P35372) | Homo sapiens | S263 | LMILRLKSVRMLSGS | 0.189 | 0.0136 | - | - | - | - | - | - | - |
| PPP1R9B (D3DTX6) | Homo sapiens | S116 | ALLKLGTSVSERVSR | - | 0.5176 | - | - | - | - | - | - | - |
| PPP1R9B (D3DTX6) | Homo sapiens | S100 | LSLPRASSLNENVDH | - | 0.6136 | - | - | - | - | - | - | - |
| CAMK2G (Q13555) | Homo sapiens | T287 | SMMHRQETVECLRKF | 0.798 | 0.3667 | - | - | - | - | - | - | - |
| VPS72 (Q15906) | Homo sapiens | T86 | RRKRRVVTKAYKEPL | 0.905 | 0.6136 | - | - | - | - | - | - | - |
| VPS72 (Q15906) | Homo sapiens | T137 | RKSMRQSTAEHTRQT | 0.990 | 0.7718 | - | - | - | - | - | - | - |
| VPS72 (Q15906) | Homo sapiens | T168 | PHCERPLTQEELLRE | 0.927 | 0.6427 | - | - | - | - | - | - | - |
| PLN (P26678) | Homo sapiens | T17 | SAIRRASTIEMPQQA | 0.522 | 0.1671 | - | - | - | - | - | - | - |
| (Q53H78) | Homo sapiens | T287 | SMMYRQETVECLKKF | - | 0.2645 | - | - | - | - | - | - | - |
| CAMK2A (Q7LDD5) | Homo sapiens | T286 | SCMHRQETVDCLKKF | - | 0.2752 | - | - | - | - | - | - | - |
Help Instructions
- Hovering on the "*" in "Substrate" column will display the text if there is a note by the annotator for the given site.
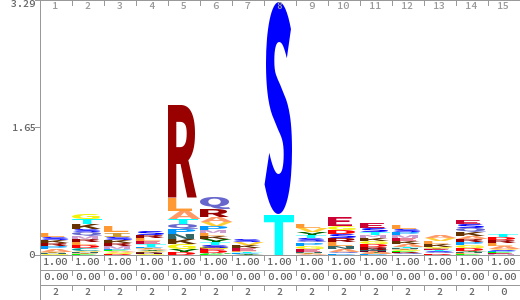
- The column having "Sequence" motif represents phosphosite and its neighbouring 7 residues (upstream and downstream).
- The score in "Conservation" column varies from 0 to 1; Higher the score more conserved is the site.
- The score in "Disorder" column varies from 0 to 1; Score higher than 0.4 suggests disorder.
- In the "Curator Assessment" column, TP refers to True Positive; FP refers to False Positive; and U refers to unknown if the annoatator cannot decide the status based on available information.
- Hovering on the number in the "Evidence Logic" column will show the name of method used to characterize/infer the phosphosite (for more details plase follow the link)