Help Instruction
- Double clicking on the substrate will redirect to the uniprot entry.
- Clicking in the block having the substrate will zoom into its phophosites
- Clicking the center of the circle will lead to zooming out.
- Clicking on the phosphosite will render the phosphosite in its 3D structure (if structure information/mapping is available).
- The color of every substrate has ben kept different in the Sunburst diagram for visual clarity.
- Clicking in the top right corner on the camara icon will download the image.
| Substrate | Organism | PSite | Sequence(+/-7) | Conservation | Disorder | Curator Assessment | Reliability | Evidence Class | Evidence Logic | PubMed | Phospho-ELM | PhosphoSite-Plus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E2F3 (O00716-1) | Homo sapiens | S124 | PALGRGGSGGGGGPP | 0.130 | 0.9106 | TP | certain | experimental | 72 | 9 | 73 | 55 | | 19917728 | - | - |
| CDC25A (P30304) | Homo sapiens | S124 | PALKRSHSDSLDHDI | 0.462 | 0.4476 | - | - | - | - | - | Non high throughput (11298456) |
|
| CHEK2 (O96017) | Homo sapiens | T383 | GETSLMRTLCGTPTY | 0.910 | 0.1162 | - | - | - | - | - | Non high throughput (11390408) |
|
| CHEK2 (O96017) | Homo sapiens | T387 | LMRTLCGTPTYLAPE | 0.857 | 0.1115 | - | - | - | - | - | Non high throughput (11390408) |
|
| CHEK2 (O96017) | Homo sapiens | T68 | SSLETVSTQELYSIP | 0.781 | 0.5577 | - | - | - | - | - | Non high throughput (11901158) |
|
| CDC25A (P30304) | Homo sapiens | S178 | LFTQRQNSAPARMLS | 0.959 | 0.6755 | - | - | - | - | - | Non high throughput (12676583) |
|
| CDC25A (P30304) | Homo sapiens | S279 | VLKRPERSQEESPPG | 0.549 | 0.8872 | - | - | - | - | - | Non high throughput (12676583) |
|
| CDC25A (P30304) | Homo sapiens | S293 | GSTKRRKSMSGASPK | 0.631 | 0.8713 | - | - | - | - | - | Non high throughput (12676583) |
|
| E2F1 (Q01094) | Homo sapiens | S364 | PLLSRMGSLRAPVDE | 0.364 | 0.6322 | - | - | - | - | - | Non high throughput (12717439) |
|
| CHEK2 (O96017) | Homo sapiens | S516 | PQVLAQPSTSRKRPR | 0.537 | 0.6661 | - | - | - | - | - | Non high throughput (12805407) |
|
| (P79923) | Xenopus laevis | S137 | PAFKRNLSYSLECEV | - | 0.2436 | - | - | - | - | - | Non high throughput (15272308) | - |
| (P79923) | Xenopus laevis | S190 | GLVQRQKSAPACMLS | - | 0.5665 | - | - | - | - | - | Non high throughput (15272308) | - |
| (P79923) | Xenopus laevis | S295 | EKSKRRKSMSEIVNE | - | 0.6375 | - | - | - | - | - | Non high throughput (15272308) | - |
| Cds1 (Q98TW0) | Xenopus laevis | T355 | GETSLMRTLCGTPTY | - | 0.1206 | - | - | - | - | - | Non high throughput (15509799) | - |
| Cds1 (Q98TW0) | Xenopus laevis | T359 | LMRTLCGTPTYLAPE | - | 0.1791 | - | - | - | - | - | Non high throughput (15509799) | - |
| TP53 (P04637) | Homo sapiens | S20 | PLSQETFSDLWKLLP | 0.567 | 0.6183 | - | - | - | - | - | Non high throughput (17339337) |
|
| CDC25C (P30307) | Homo sapiens | S216 | SGLYRSPSMPENLNR | 0.868 | 0.6136 | - | - | - | - | - | Non high throughput (10744722, 9836640) |
|
| BRCA1 (P38398) | Homo sapiens | S988 | PPLFPIKSFVKTKCK | 0.798 | 0.2645 | - | - | - | - | - | Non high throughput (12111733, 14701743) |
|
| XRCC1 (P18887) | Homo sapiens | T284 | APTRTPATAPVPARA | 0.433 | 0.8343 | - | - | - | - | - | - |
|
| E2F3 (O00716) | Homo sapiens | S124 | PALGRGGSGGGGGPP | 0.130 | 0.9106 | - | - | - | - | - | - |
|
| MDM4 (O15151) | Homo sapiens | S342 | SKLTHSLSTSDITAI | 0.727 | 0.4292 | - | - | - | - | - | - |
|
| MDM4 (O15151) | Homo sapiens | S367 | PDCRRTISAPVVRPK | 0.922 | 0.433 | - | - | - | - | - | - |
|
| REV3L (O60673) | Homo sapiens | S1075 | ENIKRTLSFRKKRSH | 0.283 | 0.5254 | - | - | - | - | - | - |
|
| CHEK2 (O96017) | Homo sapiens | S120 | YWFGRDKSCEYCFDE | 0.327 | 0.1702 | - | - | - | - | - | - |
|
| CHEK2 (O96017) | Homo sapiens | S140 | TDKYRTYSKKHFRIF | 0.969 | 0.2034 | - | - | - | - | - | - |
|
| CHEK2 (O96017) | Homo sapiens | S19 | SHGSSACSQPHGSVT | 0.500 | 0.7982 | - | - | - | - | - | - |
|
| CHEK2 (O96017) | Homo sapiens | S260 | KRKFAIGSAREADPA | 0.148 | 0.1184 | - | - | - | - | - | - |
|
| CHEK2 (O96017) | Homo sapiens | S33 | TQSQGSSSQSQGISS | 0.500 | 0.7672 | - | - | - | - | - | - |
|
| CHEK2 (O96017) | Homo sapiens | S35 | SQGSSSQSQGISSSS | 0.500 | 0.7595 | - | - | - | - | - | - |
|
| CHEK2 (O96017) | Homo sapiens | S379 | SKILGETSLMRTLCG | 0.459 | 0.1501 | - | - | - | - | - | - |
|
| CHEK2 (O96017) | Homo sapiens | S435 | SEHRTQVSLKDQITS | 0.355 | 0.4292 | - | - | - | - | - | - |
|
| MBP (P02687) | Bos taurus | S150 | HDAQGTLSKIFKLGG | - | 0.6136 | - | - | - | - | - | - |
|
| MBP (P02687) | Bos taurus | S17 | SKYLASASTMDHARH | - | 0.5901 | - | - | - | - | - | - |
|
| TP53 (P04637) | Homo sapiens | S15 | PSVEPPLSQETFSDL | 0.830 | 0.708 | - | - | - | - | - | - |
|
| TP53 (P04637) | Homo sapiens | S366 | PGGSRAHSSHLKSKK | 0.367 | 0.9081 | - | - | - | - | - | - |
|
| TP53 (P04637) | Homo sapiens | S37 | NVLSPLPSQAMDDLM | 0.107 | 0.708 | - | - | - | - | - | - |
|
| TP53 (P04637) | Homo sapiens | S378 | SKKGQSTSRHKKLMF | 0.826 | 0.7881 | - | - | - | - | - | - |
|
| RB1 (P06400) | Homo sapiens | S612 | MYLSPVRSPKKKGST | 0.442 | 0.433 | - | - | - | - | - | - |
|
| MAPT (P10636-8) | Homo sapiens | S262 | NVKSKIGSTENLKHQ | 0.606 | 0.8125 | - | - | - | - | - | - |
|
| HIST1H1C (P16403) | Homo sapiens | S36 | GGTPRKASGPPVSEL | 0.768 | 0.7415 | - | - | - | - | - | - |
|
| HIST1H1C (P16403) | Homo sapiens | S55 | VAASKERSGVSLAAL | 0.198 | 0.5296 | - | - | - | - | - | - |
|
| HIST1H1C (P16403) | Homo sapiens | S89 | LGLKSLVSKGTLVQT | 0.092 | 0.5017 | - | - | - | - | - | - |
|
| XRCC1 (P18887) | Homo sapiens | S184 | EEDESANSLRPGALF | 0.514 | 0.562 | - | - | - | - | - | - |
|
| XRCC1 (P18887) | Homo sapiens | S210 | ASDPAGPSYAAATLQ | 0.903 | 0.725 | - | - | - | - | - | - |
|
| PML (P29590-3) | Homo sapiens | S117 | ESLQRRLSVYRQIVD | 0.681 | 0.1373 | - | - | - | - | - | - |
|
| VHL (P40337) | Homo sapiens | S111 | GTGRRIHSYRGHLWL | 0.156 | 0.3249 | - | - | - | - | - | - |
|
| FOXM1 (Q08050-2) | Homo sapiens | S361 | PLLPRVSSYLVPIQF | 0.213 | 0.1671 | - | - | - | - | - | - |
|
| TRIM28 (Q13263) | Homo sapiens | S473 | SGVKRSRSGEGEVSG | 0.459 | 0.7163 | - | - | - | - | - | - |
|
| ELAVL1 (Q15717) | Homo sapiens | S100 | VSYARPSSEVIKDAN | 0.601 | 0.1914 | - | - | - | - | - | - |
|
| ELAVL1 (Q15717) | Homo sapiens | S88 | LNGLRLQSKTIKVSY | 0.093 | 0.2918 | - | - | - | - | - | - |
|
| LATS2 (Q9NRM7) | Homo sapiens | S172 | TPVTRRPSFEGTGDS | 0.526 | 0.6183 | - | - | - | - | - | - |
|
| LATS2 (Q9NRM7) | Homo sapiens | S380 | ATLARRDSLQKPGLE | 0.706 | 0.7547 | - | - | - | - | - | - |
|
| LATS2 (Q9NRM7) | Homo sapiens | S408 | PVPSRTNSFNSHQPR | 0.893 | 0.9013 | - | - | - | - | - | - |
|
| LATS2 (Q9NRM7) | Homo sapiens | S446 | HILHPVKSVRVLRPE | 0.824 | 0.6183 | - | - | - | - | - | - |
|
| AATF (Q9NY61) | Homo sapiens | S143 | SKKSRSHSAKTPGFS | 0.131 | 0.6906 | - | - | - | - | - | - |
|
| AATF (Q9NY61) | Homo sapiens | S477 | ELIERKTSSLDPNDQ | 0.237 | 0.4186 | - | - | - | - | - | - |
|
| AATF (Q9NY61) | Homo sapiens | S510 | KKVDRKASKGRKLRF | 0.618 | 0.3566 | - | - | - | - | - | - |
|
| CHEK2 (O96017) | Homo sapiens | T225 | DEYIMSKTLGSGACG | 0.135 | 0.1611 | - | - | - | - | - | - |
|
| CHEK2 (O96017) | Homo sapiens | T432 | PPFSEHRTQVSLKDQ | 0.203 | 0.4292 | - | - | - | - | - | - |
|
| MBP (P02687) | Bos taurus | T148 | KGHDAQGTLSKIFKL | - | 0.6183 | - | - | - | - | - | - |
|
| MBP (P02687) | Bos taurus | T18 | KYLASASTMDHARHG | - | 0.6322 | - | - | - | - | - | - |
|
| MBP (P02687) | Bos taurus | T33 | FLPRHRDTGILDSLG | - | 0.4619 | - | - | - | - | - | - |
|
| MBP (P02687) | Bos taurus | T64 | DGHHAARTTHYGSLP | - | 0.8125 | - | - | - | - | - | - |
|
| TP53 (P04637) | Homo sapiens | T18 | EPPLSQETFSDLWKL | 0.691 | 0.6183 | - | - | - | - | - | - |
|
| HIST1H1C (P16403) | Homo sapiens | T167 | KPAAATVTKKVAKSP | 0.099 | 0.7629 | - | - | - | - | - | - |
|
| HIST1H1C (P16403) | Homo sapiens | T92 | KSLVSKGTLVQTKGT | 0.371 | 0.5139 | - | - | - | - | - | - |
|
| TTK (P33981) | Homo sapiens | T288 | SPDCDVKTDDSVVPC | 0.231 | 0.3392 | - | - | - | - | - | - |
|
| ELAVL1 (Q15717) | Homo sapiens | T118 | SGLPRTMTQKDVEDM | 0.405 | 0.3149 | - | - | - | - | - | - |
|
| SIAH2 (O43255) | Homo sapiens | S68 | GGGAGPVSPQHHELT | 0.066 | 0.6136 | - | - | - | - | - | - |
|
| BAIAP2L1 (Q9UHR4) | Homo sapiens | S331 | PSLQRSVSVATGLNM | 0.970 | 0.5951 | - | - | - | - | - | - |
|
| PSME3 (P61289) | Homo sapiens | S247 | EKIKRPRSSNAETLY | 0.675 | 0.3948 | - | - | - | - | - | - |
|
| CDK11B (P21127) | Homo sapiens | S752 | QRVKRGTSPRPPEGG | 0.705 | 0.8085 | - | - | - | - | - | - |
|
| SIAH2 (O43255) | Homo sapiens | S28 | PQPQHTPSPAAPPAA | 0.500 | 0.8741 | - | - | - | - | - | - |
|
| SIAH2 (O43255) | Homo sapiens | T26 | PPPQPQHTPSPAAPP | 0.500 | 0.9168 | - | - | - | - | - | - |
|
| SIAH2 (O43255) | Homo sapiens | T119 | PTCRGALTPSIRNLA | 0.052 | 0.0967 | - | - | - | - | - | - |
|
| BLM (P54132) | Homo sapiens | T182 | SHFVRVSTAQKSKKG | 0.470 | 0.708 | - | - | - | - | - | - |
|
| BDNF (P23560) | Homo sapiens | T62 | GLTSLADTFEHVIEE | 0.708 | 0.3019 | - | - | - | - | - | - |
|
| MBP (P02686-3) | Homo sapiens | S178 | VDAQGTLSKIFKLGG | 0.870 | 0.4619 | - | - | - | - | - | - | - |
| CHEK2 (O96017-12) | Homo sapiens | S487 | PQVLAQPSTSRKRPR | 0.351 | 0.6661 | - | - | - | - | - | - | - |
| CHEK2 (O96017-9) | Homo sapiens | S559 | PQVLAQPSTSRKRPR | - | 0.6661 | - | - | - | - | - | - | - |
| PML (P29590) | Homo sapiens | S117 | ESLQRRLSVYRQIVD | 0.681 | 0.1373 | - | - | - | - | - | - | - |
| FOXM1 (Q08050) | Homo sapiens | S376 | PLLPRVSSYLVPIQF | 0.844 | 0.2129 | - | - | - | - | - | - | - |
| CDK11A (Q9UQ88) | Homo sapiens | S740 | QRVKRGTSPRPPEGG | 0.835 | 0.8013 | - | - | - | - | - | - | - |
| CDC25A (P30304) | Homo sapiens | S298 | RKSMSGASPKESTNP | 0.371 | 0.8375 | - | - | - | - | - | - | - |
| CHEK2 (O96017-4) | Homo sapiens | T292 | GETSLMRTLCGTPTY | 0.246 | 0.1162 | - | - | - | - | - | - | - |
| CHEK2 (O96017-12) | Homo sapiens | T68 | SSLETVSTQELYSIP | 0.781 | 0.5577 | - | - | - | - | - | - | - |
| BRCA2 (P51587) | Homo sapiens | T3387 | LRLKRRCTTSLIKEQ | 0.831 | 0.4119 | - | - | - | - | - | - | - |
| MBP (P02686) | Homo sapiens | T21 | KASTNSETNRGESEK | 0.500 | 0.9168 | - | - | - | - | - | - | - |
Help Instructions
- Hovering on the "*" in "Substrate" column will display the text if there is a note by the annotator for the given site.
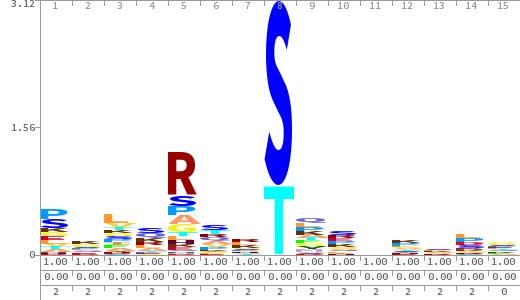
- The column having "Sequence" motif represents phosphosite and its neighbouring 7 residues (upstream and downstream).
- The score in "Conservation" column varies from 0 to 1; Higher the score more conserved is the site.
- The score in "Disorder" column varies from 0 to 1; Score higher than 0.4 suggests disorder.
- In the "Curator Assessment" column, TP refers to True Positive; FP refers to False Positive; and U refers to unknown if the annoatator cannot decide the status based on available information.
- Hovering on the number in the "Evidence Logic" column will show the name of method used to characterize/infer the phosphosite (for more details plase follow the link)