Help Instruction
- Double clicking on the substrate will redirect to the uniprot entry.
- Clicking in the block having the substrate will zoom into its phophosites
- Clicking the center of the circle will lead to zooming out.
- Clicking on the phosphosite will render the phosphosite in its 3D structure (if structure information/mapping is available).
- The color of every substrate has ben kept different in the Sunburst diagram for visual clarity.
- Clicking in the top right corner on the camara icon will download the image.
| Substrate | Organism | PSite | Sequence(+/-7) | Conservation | Disorder | Curator Assessment | Reliability | Evidence Class | Evidence Logic | PubMed | Phospho-ELM | PhosphoSite-Plus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dnm1l (O35303) | Rattus norvegicus | S656 | VPVARKLSAREQRDC | - | 0.4119 | TP | certain | experimental | 72 | 73 | 8 | 55 | | 18695047 | - |
|
| EIF4G3 (O43432) | Homo sapiens | S1156 | NTFMRGGSSKDLLDN | 0.511 | 0.7459 | TP | certain | experimental | 72 | 73 | 8 | 107 | | 14507913 | - |
|
| Hspb1 (P14602) | Mus musculus | S15 | FSLLRSPSWEPFRDW | - | 0.1092 | TP | likely | experimental | 73 | 214 | | 12538590 | - |
|
| RPS19 * (P39019 ) | Homo sapiens | S59 | WFYTRAASTARHLYL | 0.491 | 0.2715 | - | - | - | - | - | - |
|
| Hdac7 (Q8C2B3) | Mus musculus | S178 | FPLRKTVSEPNLKLR | - | 0.6906 | TP | certain | experimental | 72 | 74 | 8 | 55 | | 16956611 | - |
|
| Rps19 (Q9CZX8) | Mus musculus | S59 | WFYTRAASTARHLYL | - | 0.2783 | TP | certain | experimental | 72 | 73 | 55 | 62 | | 19200342 | - |
|
| HDAC9 * (Q9UKV0 ) | Homo sapiens | S220 | FPLRKTASEPNLKVR | 0.893 | 0.6806 | U | likely | experimental | 72 | 8 | 73 | | 12202037 | - |
|
| HDAC9 * (Q9UKV0 ) | Homo sapiens | S451 | RPLNRTQSAPLPQST | 0.938 | 0.892 | U | likely | experimental | 72 | 8 | 73 | | 12202037 | - |
|
| CAMK1 (Q14012) | Homo sapiens | T177 | DPGSVLSTACGTPGY | 0.726 | 0.3215 | TP | certain | experimental | 8 | 73 | | 7641687 | Non high throughput (7641687, 8253780) | - |
| Snf1lk2 (Q3LRT3) | Rattus norvegicus | T484 | RSGQRRHTLSEVTNQ | - | 0.6531 | - | - | - | - | - | - |
|
| Sik1 (Q9R1U5) | Rattus norvegicus | T322 | LGIDRQRTVESLQNS | - | 0.4051 | TP | certain | experimental | 73 | 8 | | 17939993 | - |
|
| Rps19 (P17074) | Rattus norvegicus | S59 | WFYTRAASTARHLYL | - | 0.2783 | TP | certain | experimental | 9 | 216 | 148 | 72 | 73 | | 19200342 | - | - |
| Mark2 (O08679) | Rattus norvegicus | S91 | IDKTQLNSSSLQKLF | - | 0.1914 | TP | certain | experimental | 66 | 3 | 23 | 9 | 45 | | 17442826 | - | - |
| Mark2 (O08679) | Rattus norvegicus | S92 | DKTQLNSSSLQKLFR | - | 0.2541 | TP | certain | experimental | 66 | 3 | 23 | 9 | 45 | | 17442826 | - | - |
| Mark2 (O08679) | Rattus norvegicus | S93 | KTQLNSSSLQKLFRE | - | 0.1731 | TP | certain | experimental | 66 | 3 | 23 | 9 | 45 | | 17442826 | - | - |
| Hdac7 (Q8C2B3) | Mus musculus | S344 | RPLNRTRSEPLPPSA | - | 0.892 | TP | certain | experimental | 9 | 55 | 216 | | 16956611 | - | - |
| Hdac7 (Q8C2B3) | Mus musculus | S479 | RPLSRTQSSPAAPVS | - | 0.6851 | TP | certain | experimental | 9 | 55 | 216 | | 16956611 | - | - |
| HDAC5 (Q9UQL6) | Homo sapiens | S259 | FPLRKTASEPNLKVR | 0.835 | 0.6712 | TP | certain | experimental | 9 | 143 | | 11081517 | - | - |
| HDAC5 (Q9UQL6) | Homo sapiens | S498 | RPLSRTQSSPLPQSP | 0.940 | 0.9126 | TP | certain | experimental | 9 | 143 | | 11081517 | - | - |
| GCM1 (Q9NP62) | Homo sapiens | S47 | YAKHIYSSEDKNAQR | 0.505 | 0.4476 | TP | certain | experimental | 72 | 73 | 214 | 62 | 55 | | 21791615 | - | - |
| Numb (Q2LC84) | Rattus norvegicus | S275 | EQLARQGSFRGFPAL | - | 0.5493 | TP | certain | experimental | 72 | 73 | 55 | 8 | 214 | | 17022975 | - | - |
| Numb (Q2LC84) | Rattus norvegicus | S294 | SPFKRQLSLRINELP | - | 0.5807 | TP | certain | experimental | 72 | 73 | 55 | 8 | | 17022975 | - | - |
| CAMK2A (Q9UQM7-2) | Homo sapiens | S332 | DGVKKRKSSSSVQLM | 0.087 | 0.5854 | TP | certain | experimental | 8 | 73 | | 9677407 | - | - |
| EP300 (Q09472) | Homo sapiens | S89 | SELLRSGSSPNLNMG | 0.391 | 0.6851 | TP | certain | - | 8 | 73 | 135 | | 12538590 | - | - |
| DNM1L (O00429-4) | Homo sapiens | S600 | VPVARKLSAREQRDC | 0.162 | 0.384 | TP | certain | experimental | 9 | 73 | 55 | 72 | | 18695047 | - | - |
| SIK2 (Q9H0K1) | Homo sapiens | T484 | RSGQRRHTLSEVTNQ | 0.896 | 0.6482 | TP | certain | experimental | 55 | 73 | 9 | | 21220102 | - | - |
| Mark2 (O08679) | Rattus norvegicus | T294 | LNPSKRGTLEQIMKD | - | 0.1702 | TP | certain | experimental | 66 | 3 | 23 | 9 | 45 | | 17442826 | - | - |
| CDKN1B (P46527) | Homo sapiens | T157 | GIRKRPATDDSSTQN | 0.142 | 0.8198 | TP | certain | experimental | 3 | 55 | 148 | 73 | 9 | 36 | | 23707388 | - | - |
| CDKN1B (P46527) | Homo sapiens | T198 | PGLRRRQT_______ | 0.507 | 0.9488 | TP | certain | experimental | 55 | 72 | 73 | 36 | 3 | 9 | | 23707388 | - | - |
| Arhgef7 (O55043) | Rattus norvegicus | S516 | RKPERKPSDEEFAVR | - | 0.7672 | TP | certain | experimental | 72 | 73 | 9 | 55 | 23 | | 18184567 | - | - |
| KRT18 (P05783) | Homo sapiens | S53 | ISVSRSTSFRGGMGS | 0.418 | 0.4476 | - | - | - | - | - | Non high throughput (7523419) | - |
| CAMK4 (Q16566) | Homo sapiens | T200 | EHQVLMKTVCGTPGY | 0.785 | 0.087 | - | - | - | - | - | Non high throughput (15143065, 8702940) | - |
| Th (P04177) | Rattus norvegicus | S19 | KGFRRAVSEQDAKQA | - | 0.6851 | - | - | - | - | - | - |
|
| Th (P04177) | Rattus norvegicus | S40 | RFIGRRQSLIEDARK | - | 0.5854 | - | - | - | - | - | - |
|
| RYR1 (P11716) | Oryctolagus cuniculus | S2843 | KKKTRKISQTAQTYD | - | 0.6906 | - | - | - | - | - | - |
|
| MYL12A (P19105) | Homo sapiens | S19 | KRPQRATSNVFAMFD | 0.788 | 0.5577 | - | - | - | - | - | - | - |
| MYL9 (P24844) | Homo sapiens | S20 | KRPQRATSNVFAMFD | 0.877 | 0.5577 | - | - | - | - | - | - |
|
| LIPE (Q05469) | Homo sapiens | S855 | EPMRRSVSEAALAQP | 0.756 | 0.7415 | - | - | - | - | - | - |
|
| Pklr (P12928) | Rattus norvegicus | T48 | RASVAQLTQELGTAF | - | 0.1554 | - | - | - | - | - | - |
|
| Pklr (P12928) | Rattus norvegicus | T53 | QLTQELGTAFFQQQQ | - | 0.2988 | - | - | - | - | - | - |
|
| Pklr (P12928-2) | Rattus norvegicus | T17 | RASVAQLTQELGTAF | - | 0.1611 | - | - | - | - | - | - |
|
| Pklr (P12928-2) | Rattus norvegicus | T22 | QLTQELGTAFFQQQQ | - | 0.3019 | - | - | - | - | - | - |
|
| Cdkn1b (P46414) | Mus musculus | T170 | QNKRANRTEENVSDG | - | 0.9329 | - | - | - | - | - | - |
|
| Cdkn1b (P46414) | Mus musculus | T197 | KPGLRRQT_______ | - | 0.9473 | - | - | - | - | - | - |
|
| ARHGEF7 (Q14155-1) | Homo sapiens | S516 | RKPERKPSDEEFASR | 0.344 | 0.8235 | - | - | - | - | - | - | - |
| DNM1L (O00429) | Homo sapiens | S637 | VPVARKLSAREQRDC | 0.300 | 0.4119 | - | - | - | - | - | - | - |
| CREB1 (P16220) | Homo sapiens | S133 | EILSRRPSYRKILND | - | 0.4979 | - | - | - | - | - | - | - |
| SYN1 (P17600) | Homo sapiens | S9 | NYLRRRLSDSNFMAN | 0.991 | 0.5533 | - | - | - | - | - | - | - |
| ATF1 (P18846) | Homo sapiens | S63 | GILARRPSYRKILKD | 0.920 | 0.4864 | - | - | - | - | - | - | - |
| NOS1 (P29475) | Homo sapiens | S746 | LAEAVKFSAKLMGQA | 0.504 | 0.1583 | - | - | - | - | - | - | - |
| NUMB (P49757) | Homo sapiens | S276 | EQLARQGSFRGFPAL | 0.857 | 0.5577 | - | - | - | - | - | - | - |
| NUMB (P49757) | Homo sapiens | S295 | SPFKRQLSLRINELP | 0.825 | 0.5807 | - | - | - | - | - | - | - |
| ARHGEF7 (Q14155) | Homo sapiens | S694 | RKPERKPSDEEFASR | 0.809 | 0.8198 | - | - | - | - | - | - | - |
| MARK2 (Q7KZI7) | Homo sapiens | S91 | IDKTQLNSSSLQKLF | 0.008 | 0.1881 | - | - | - | - | - | - | - |
| MARK2 (Q7KZI7) | Homo sapiens | S92 | DKTQLNSSSLQKLFR | 0.186 | 0.2541 | - | - | - | - | - | - | - |
| MARK2 (Q7KZI7) | Homo sapiens | S93 | KTQLNSSSLQKLFRE | 0.653 | 0.1731 | - | - | - | - | - | - | - |
| HDAC5 (Q9UQL6-3) | Homo sapiens | S260 | FPLRKTASEPNLKVR | 0.838 | 0.6712 | - | - | - | - | - | - | - |
| HDAC5 (Q9UQL6-3) | Homo sapiens | S499 | RPLSRTQSSPLPQSP | 0.120 | 0.9126 | - | - | - | - | - | - | - |
| TH (P07101) | Homo sapiens | S19 | KGFRRAVSELDAKQA | 0.455 | 0.5665 | - | - | - | - | - | - | - |
| TH (P07101) | Homo sapiens | S44 | PGPSLTGSPWPGTAA | 0.066 | 0.8198 | - | - | - | - | - | - | - |
| HSPB1 (P04792) | Homo sapiens | S15 | FSLLRGPSWDPFRDW | 0.564 | 0.1323 | - | - | - | - | - | - | - |
| RYR1 (P21817) | Homo sapiens | S2843 | KKKTRKISQSAQTYD | 0.703 | 0.6851 | - | - | - | - | - | - | - |
| RPS19 (B0ZBD0) | Homo sapiens | S59 | WFYTRAASTARHLYL | - | 0.2715 | - | - | - | - | - | - | - |
| HDAC7 (Q8WUI4) | Homo sapiens | S508 | SQARVLSSSETPART | 0.206 | 0.6531 | - | - | - | - | - | - | - |
| NOS1 (B3VK56) | Homo sapiens | S852 | SYKVRFNSVSSYSDS | - | 0.346 | - | - | - | - | - | - | - |
| DNM1L (O00429-3) | Homo sapiens | S639 | VRKNIQDSVPKAVMH | 0.552 | 0.0851 | - | - | - | - | - | - | - |
| HDAC7 (Q8WUI4-6) | Homo sapiens | S177 | FPLRKTVSEPNLKLR | 0.366 | 0.6227 | - | - | - | - | - | - | - |
| SIK1 (P57059) | Homo sapiens | T322 | LGVDRQRTVESLQNS | 0.753 | 0.4507 | - | - | - | - | - | - | - |
| MARK2 (Q7KZI7) | Homo sapiens | T294 | LNPSKRGTLEQIMKD | 0.010 | 0.1702 | - | - | - | - | - | - | - |
| PKLR (P30613) | Homo sapiens | T48 | RASVAQLTQELGTAF | 0.038 | 0.1373 | - | - | - | - | - | - | - |
| PKLR (P30613) | Homo sapiens | T53 | QLTQELGTAFFQQQQ | 0.045 | 0.268 | - | - | - | - | - | - | - |
| (Q16716) | Homo sapiens | T40 | RASVAQLTQELGTAF | - | 0.2064 | - | - | - | - | - | - | - |
| (Q16716) | Homo sapiens | T45 | QLTQELGTAFFQQQQ | - | 0.346 | - | - | - | - | - | - | - |
| SNF1LK2 (A0A024R3G7) | Homo sapiens | T484 | RSGQRRHTLSEVTNQ | - | 0.6482 | - | - | - | - | - | - | - |
| SIK1B (A0A0B4J2F2) | Homo sapiens | T322 | LGVDRQRTVESLQNS | - | 0.4507 | - | - | - | - | - | - | - |
| CDKN1B (P46527) | Homo sapiens | T170 | QNKRANRTEENVSDG | 0.565 | 0.9346 | - | - | - | - | - | - | - |
Help Instructions
- Hovering on the "*" in "Substrate" column will display the text if there is a note by the annotator for the given site.
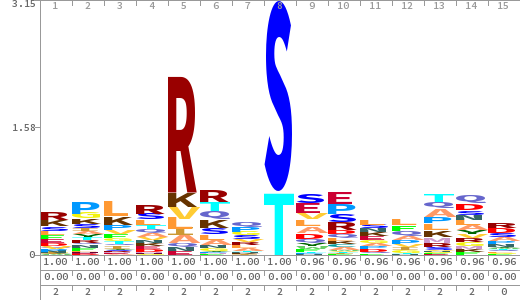
- The column having "Sequence" motif represents phosphosite and its neighbouring 7 residues (upstream and downstream).
- The score in "Conservation" column varies from 0 to 1; Higher the score more conserved is the site.
- The score in "Disorder" column varies from 0 to 1; Score higher than 0.4 suggests disorder.
- In the "Curator Assessment" column, TP refers to True Positive; FP refers to False Positive; and U refers to unknown if the annoatator cannot decide the status based on available information.
- Hovering on the number in the "Evidence Logic" column will show the name of method used to characterize/infer the phosphosite (for more details plase follow the link)