Help Instruction
- Double clicking on the substrate will redirect to the uniprot entry.
- Clicking in the block having the substrate will zoom into its phophosites
- Clicking the center of the circle will lead to zooming out.
- Clicking on the phosphosite will render the phosphosite in its 3D structure (if structure information/mapping is available).
- The color of every substrate has ben kept different in the Sunburst diagram for visual clarity.
- Clicking in the top right corner on the camara icon will download the image.
| Substrate | Organism | PSite | Sequence(+/-7) | Conservation | Disorder | Curator Assessment | Reliability | Evidence Class | Evidence Logic | PubMed | Phospho-ELM | PhosphoSite-Plus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FOXO3 (O43524) | Homo sapiens | S253 | APRRRAVSMDNSNKY | 0.762 | 0.6906 | TP | certain | experimental | 72 | 9 | | 18593906 | - |
|
| FOXO3 (O43524) | Homo sapiens | T32 | QSRPRSCTWPLQRPE | 0.886 | 0.8343 | TP | certain | - | 72 | 9 | | 18593906 | - |
|
| MARK3 * (P27448-3 ) | Homo sapiens | S96 | KTQLNPTSLQKLFRE | 0.741 | 0.2193 | U | likely | experimental | 214 | 9 | | 15319445 | - |
|
| MARK3 * (P27448-3 ) | Homo sapiens | T90 | AIKIIDKTQLNPTSL | 0.682 | 0.3019 | U | likely | experimental | 214 | 9 | | 15319445 | - |
|
| MARK3 * (P27448-3 ) | Homo sapiens | T95 | DKTQLNPTSLQKLFR | 0.017 | 0.3019 | U | likely | experimental | 214 | 9 | | 15319445 | - |
|
| MDM2 (Q00987) | Homo sapiens | S166 | SSRRRAISETEENSD | 0.815 | 0.7718 | TP | certain | experimental | 72 | 73 | 8 | 55 | 107 | 23 | | 18467333 | - |
|
| MDM2 * (Q00987 ) | Homo sapiens | S186 | RQRKRHKSDSISLSF | 0.958 | 0.6043 | TP | likely | experimental | 73 | 8 | 55 | 107 | 72 | 23 | | 18467333 | - |
|
| POU5F1 * (Q01860 ) | Homo sapiens | S288 | RRQKGKRSSSDYAQR | 0.512 | 0.4409 | U | likely | experimental | 73 | 214 | | 22474382 | - |
|
| POU5F1 * (Q01860 ) | Homo sapiens | S289 | RQKGKRSSSDYAQRE | 0.382 | 0.5577 | U | likely | experimental | 73 | 214 | | 22474382 | - |
|
| MAP3K5 (Q99683) | Homo sapiens | S83 | ATRGRGSSVGGGSRR | 0.816 | 0.6661 | TP | certain | experimental | 72 | 73 | 9 | 55 | | 19749799 | - | - |
| EIF4B (P23588) | Homo sapiens | S406 | RPRERHPSWRSEETQ | 0.581 | 0.8462 | TP | certain | experimental | 72 | 9 | 148 | 28 | 40 | 55 | | 24777602 | - | - |
| CDKN1A (P38936) | Homo sapiens | S146 | GRKRRQTSMTDFYHS | 0.575 | 0.7163 | TP | certain | experimental | 72 | 9 | 55 | | 17855660 | - | - |
| FLT3 (P36888) | Homo sapiens | S935 | FDSRKRPSFPNLTSF | 0.457 | 0.0568 | TP | certain | experimental | 72 | 40 | 148 | 9 | 28 | | 24040307 | - | - |
| LANA1 (Q9QR71) | Human herpesvirus 8 type P (isolate GK18) | S205 | SLRKRRLSSPQGPST | - | 0.892 | TP | likely | experimental | 72 | 62 | | 16647097 | - | - |
| LANA1 (Q9QR71) | Human herpesvirus 8 type P (isolate GK18) | S206 | LRKRRLSSPQGPSTL | - | 0.853 | TP | likely | experimental | 72 | 62 | | 16647097 | - | - |
| RELA (Q04206) | Homo sapiens | S276 | SMQLRRPSDRELSEP | 0.767 | 0.5296 | TP | certain | experimental | 72 | 9 | 55 | | 19911008 | - | - |
| Bad (Q61337) | Mus musculus | S112 | ETRSRHSSYPAGTEE | - | 0.8828 | TP | certain | experimental | 72 | 135 | 133 | 30 | 73 | 9 | | 16403219 | - | - |
| Bad (Q61337) | Mus musculus | S136 | PFRGRSRSAPPNLWA | - | 0.6661 | TP | certain | experimental | 72 | 135 | 133 | 30 | 73 | 9 | | 16403219 | - | - |
| Bad (Q61337) | Mus musculus | S155 | GRELRRMSDEFEGSF | - | 0.5456 | TP | certain | experimental | 72 | 135 | 133 | 30 | 73 | 9 | | 16403219 | - | - |
| CHEK1 (O14757) | Homo sapiens | S280 | AKRPRVTSGGVSESP | 0.246 | 0.6227 | TP | certain | experimental | 148 | 72 | 73 | 55 | 9 | | 23748345 | - | - |
| RUNX3 (Q13761) | Homo sapiens | S149 | GRSGRGKSFTLTITV | 0.497 | 0.3117 | FP | likely | experimental | 73 | 72 | 3 | 31 | 9 | 39 | 66 | | 18767071 | - | - |
| HIST1H3A (P68431) | Homo sapiens | S11 | TKQTARKSTGGKAPR | 0.711 | 0.7505 | TP | certain | experimental | 72 | 73 | 55 | 23 | 9 | | 17643117 | - | - |
| Myc (P01108) | Mus musculus | S329 | AKRAKLDSGRVLKQI | - | 0.6375 | TP | certain | experimental | 72 | 62 | 73 | 139 | | 18438430 | - | - |
| EIF4B (P23588) | Homo sapiens | S422 | RERSRTGSESSQTGT | 0.591 | 0.9457 | TP | certain | experimental | 40 | 55 | 9 | 72 | 9 | | 23749639 | - | - |
| (Q9DUM3) | Human herpesvirus 8 | S205 | SLRKRRLSSPQGPST | - | 0.9013 | TP | certain | experimental | 72 | 73 | 74 | 20 | 62 | | 19266083 | - | - |
| (Q9DUM3) | Human herpesvirus 8 | S206 | LRKRRLSSPQGPSTL | - | 0.8655 | TP | certain | experimental | 72 | 73 | 74 | 20 | 62 | | 19266083 | - | - |
| CDKN1A * (P38936 ) | Homo sapiens | T145 | QGRKRRQTSMTDFYH | 0.981 | 0.725 | TP | certain | experimental | 72 | 9 | 55 | 73 | 39 | | 17855660 | - | - |
| RUNX3 (Q13761) | Homo sapiens | T151 | SGRGKSFTLTITVFT | 0.500 | 0.4369 | FP | likely | experimental | 73 | 72 | 3 | 31 | 9 | 39 | 66 | | 18767071 | - | - |
| RUNX3 * (Q13761 ) | Homo sapiens | T153 | RGKSFTLTITVFTNP | 0.520 | 0.4476 | FP | likely | experimental | 73 | 72 | 3 | 31 | 9 | 39 | 66 | | 18767071 | - | - |
| RUNX3 (Q13761) | Homo sapiens | T155 | KSFTLTITVFTNPTQ | 0.518 | 0.4292 | FP | likely | experimental | 73 | 72 | 3 | 31 | 9 | 39 | 66 | | 18767071 | - | - |
| CDC25A (P30304) | Homo sapiens | S116 | PQKLLGCSPALKRSH | 0.618 | 0.5456 | - | - | - | - | - | Non high throughput (12411508) | - |
| CXCR4 (P61073) | Homo sapiens | S339 | GKRGGHSSVSTESES | 0.806 | 0.3392 | - | - | - | - | - | - |
|
| Cxcr4 (P70658) | Mus musculus | S346 | GKRGGHSSVSTESES | - | 0.3392 | - | - | - | - | - | - |
|
| SKP2 (Q13309) | Homo sapiens | S64 | SNLGHPESPPRKRLK | 0.183 | 0.5854 | - | - | - | - | - | - |
|
| SKP2 (Q13309) | Homo sapiens | S72 | PPRKRLKSKGSDKDF | 0.303 | 0.4801 | - | - | - | - | - | - |
|
| CDKN1B (P46527) | Homo sapiens | T157 | GIRKRPATDDSSTQN | 0.142 | 0.8198 | - | - | - | - | - | - |
|
| CDKN1B (P46527) | Homo sapiens | T198 | PGLRRRQT_______ | 0.507 | 0.9488 | - | - | - | - | - | - |
|
| SKP2 (Q13309) | Homo sapiens | T417 | WGIKCRLTLQKPSCL | 0.106 | 0.0244 | - | - | - | - | - | - |
|
| ABCG2 (Q9UNQ0) | Homo sapiens | T362 | GEKKKKITVFKEISY | 0.125 | 0.1731 | - | - | - | - | - | - |
|
| UHRF1 (Q96T88-2) | Homo sapiens | S311 | DNPMRRKSGPSCKHC | 0.134 | 0.4766 | TP | certain | experimental | 73 | 55 | 8 | 214 | 23 | 3 | 72 | | 28394343 | - |
|
| BAD (Q92934) | Homo sapiens | S75 | EIRSRHSSYPAGTED | 0.509 | 0.8198 | TP | certain | experimental | 73 | 55 | | 21187157 | - |
|
| IRS1 (P35568) | Homo sapiens | S1101 | GCRRRHSSETFSSTP | 0.923 | 0.8085 | TP | certain | experimental | 72 | 140 | 73 | 8 | 55 | 148 | | 26956053 | - |
|
| IRS2 (Q9Y4H2) | Homo sapiens | S1149 | GGRRRHSSETFSSTT | 0.742 | 0.9249 | TP | certain | experimental | 73 | 55 | | 26956053 | - |
|
| HBP1 (O60381) | Homo sapiens | S380 | MARQRRASLSCGGPG | 0.862 | 0.4685 | TP | certain | experimental | 72 | 73 | 23 | 3 | 55 | 214 | 8 | | 28348080 | - |
|
| NKX3-1 (Q99801) | Homo sapiens | S186 | TKRKQLSSELGDLEK | 0.270 | 0.5017 | TP | certain | experimental | 73 | 214 | 72 | 8 | | 23129228 | - |
|
| Notch1 (Q01705) | Mus musculus | S2152 | GKKARKPSTKGLACG | - | 0.712 | TP | certain | experimental | 73 | 8 | 148 | 214 | 55 | 72 | | 27281612 | - |
|
| NKX3-1 * (Q99801 ) | Homo sapiens | S195 | LGDLEKHSSLPALKE | 0.244 | 0.374 | TP | likely | experimental | 73 | 214 | | 23129228 | - |
|
| NKX3-1 * (Q99801 ) | Homo sapiens | S196 | GDLEKHSSLPALKEE | 0.106 | 0.3704 | TP | likely | experimental | 73 | 214 | | 23129228 | - |
|
| AR * (P10275 ) | Homo sapiens | S215 | SGRAREASGAPTSSK | 0.312 | 0.7881 | TP | certain | experimental | 3 | 23 | 73 | 72 | 55 | 8 | | 22584579 | - |
|
| HBP1 (O60381) | Homo sapiens | S372 | SAVYVLSSMARQRRA | 0.761 | 0.2752 | TP | certain | experimental | 72 | 73 | 23 | 3 | 55 | 214 | 8 | | 28348080 | - |
|
| NKX3-1 (Q99801) | Homo sapiens | S185 | KTKRKQLSSELGDLE | 0.098 | 0.5098 | TP | certain | experimental | 73 | 214 | 8 | 72 | | 23129228 | - |
|
| FOXP3 * (Q9BZS1 ) | Homo sapiens | S422 | KKRSQRPSRCSNPTP | 0.818 | 0.712 | TP | certain | experimental | 23 | 53 | 55 | 72 | 73 | | 25096571 | - |
|
| AR * (P10275 ) | Homo sapiens | T851 | ACKRKNPTSCSRRFY | 0.094 | 0.1137 | TP | certain | experimental | 3 | 23 | 73 | 72 | 55 | 8 | | 22584579 | - |
|
| NKX3-1 * (Q99801 ) | Homo sapiens | T89 | AAPEEAETLAETEPE | 0.416 | 0.9039 | TP | likely | experimental | 73 | 214 | | 23129228 | - |
|
| PIM1 (P11309-2) | Homo sapiens | S352 | FFRQRVSSECQHLIR | 0.208 | 0.0334 | - | - | - | - | - | - | - |
| FOXO1 (Q12778) | Homo sapiens | S256 | SPRRRAASMDNNSKF | 0.725 | 0.7163 | TP | certain | predicted | 14 | | 18593906 | - | - |
| FOXO1 (Q12778) | Homo sapiens | S319 | TFRPRTSSNASTISG | 0.765 | 0.5665 | TP | certain | predicted | 14 | | 18593906 | - | - |
| MYC (P01106) | Homo sapiens | S329 | AKRVKLDSVRVLRQI | 0.225 | 0.4979 | TP | certain | predicted | 14 | | 18438430 | - | - |
| Myc * (P01108 ) | Mus musculus | S329 | AKRAKLDSGRVLKQI | - | 0.6375 | TP | certain | experimental | 73 | 72 | 8 | 55 | | 18438430 | - | - |
| PIM1 (P11309) | Homo sapiens | S189 | LKLIDFGSGALLKDT | 0.122 | 0.0592 | - | - | - | - | - | - | - |
| RP9 * (Q8TA86 ) | Homo sapiens | S212 | KKRKHKSSKSNEGSD | 0.669 | 0.8792 | - | - | - | - | - | - | - |
| RP9 (Q8TA86) | Homo sapiens | S214 | RKHKSSKSNEGSDSE | 0.270 | 0.8792 | - | - | - | - | - | - | - |
| BAD (Q92934) | Homo sapiens | S118 | GRELRRMSDEFVDSF | 0.787 | 0.5758 | - | - | - | - | - | - | - |
| BAD (Q92934) | Homo sapiens | S99 | PFRGRSRSAPPNLWA | 0.900 | 0.7982 | - | - | - | - | - | - | - |
| (B4DZQ9) | Homo sapiens | S75 | EIRSRHSSYPAGTED | - | 0.853 | - | - | - | - | - | - | - |
| (B4DZQ9) | Homo sapiens | S99 | PFRGRSRSAPPNLWA | - | 0.8343 | - | - | - | - | - | - | - |
| (B4DZQ9) | Homo sapiens | S118 | GRELRRMSDEFVDSF | - | 0.6427 | - | - | - | - | - | - | - |
| CDC25A (A0A024R2S4) | Homo sapiens | S116 | PQKLLGCSPALKRSH | - | 0.5456 | - | - | - | - | - | - | - |
| NOTCH1 (P46531) | Homo sapiens | S2162 | GKKVRKPSSKGLACG | 0.565 | 0.6269 | - | - | - | - | - | - | - |
| FOXO1 (Q12778) | Homo sapiens | T24 | LPRPRSCTWPLPRPE | 0.776 | 0.6183 | TP | certain | predicted | 14 | | 18593906 | - | - |
| MARK3 * (P27448 ) | Homo sapiens | T95 | DKTQLNPTSLQKLFR | 0.017 | 0.3019 | FP | unlikely | experimental | 73 | 214 | | 15319445 | - | - |
| AKT1S1 (Q96B36) | Homo sapiens | T246 | LPRPRLNTSDFQKLK | 0.738 | 0.5098 | TP | certain | experimental | 73 | 36 | 55 | 148 | | 19276681 | - | - |
Help Instructions
- Hovering on the "*" in "Substrate" column will display the text if there is a note by the annotator for the given site.
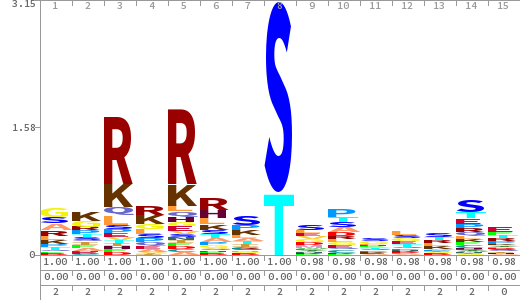
- The column having "Sequence" motif represents phosphosite and its neighbouring 7 residues (upstream and downstream).
- The score in "Conservation" column varies from 0 to 1; Higher the score more conserved is the site.
- The score in "Disorder" column varies from 0 to 1; Score higher than 0.4 suggests disorder.
- In the "Curator Assessment" column, TP refers to True Positive; FP refers to False Positive; and U refers to unknown if the annoatator cannot decide the status based on available information.
- Hovering on the number in the "Evidence Logic" column will show the name of method used to characterize/infer the phosphosite (for more details plase follow the link)