Help Instruction
- Double clicking on the substrate will redirect to the uniprot entry.
- Clicking in the block having the substrate will zoom into its phophosites
- Clicking the center of the circle will lead to zooming out.
- Clicking on the phosphosite will render the phosphosite in its 3D structure (if structure information/mapping is available).
- The color of every substrate has ben kept different in the Sunburst diagram for visual clarity.
- Clicking in the top right corner on the camara icon will download the image.
| Substrate | Organism | PSite | Sequence(+/-7) | Conservation | Disorder | Curator Assessment | Reliability | Evidence Class | Evidence Logic | PubMed | Phospho-ELM | PhosphoSite-Plus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TFAP2A (P05549) | Homo sapiens | S258 | GVLRRAKSKNGGRSL | 0.747 | 0.4292 | TP | certain | experimental | 72 | 9 | 55 | 74 | | 18845787 | - | - |
| TFAP2A (P05549) | Homo sapiens | S326 | EFLNRQHSDPNEQVT | 0.153 | 0.5758 | TP | certain | experimental | 72 | 9 | 55 | 74 | | 18845787 | - | - |
| COL4A3BP (Q9Y5P4) | Homo sapiens | S132 | SSLRRHGSMVSLVSG | 0.931 | 0.4441 | TP | certain | experimental | 62 | 55 | 72 | 73 | | 17591919 | - | - |
| CTTN (Q14247) | Homo sapiens | S298 | EKLAKHESQQDYSKG | 0.484 | 0.5456 | TP | certain | experimental | 72 | 73 | 55 | 9 | | 20363754 | - | - |
| CREB1 (P16220) | Homo sapiens | S98 | KDLKRLFSGTQISTI | - | 0.4186 | TP | certain | experimental | 62 | 72 | 55 | | 17389598 | - | - |
| CREB1 (P16220) | Homo sapiens | S133 | EILSRRPSYRKILND | - | 0.4979 | TP | certain | experimental | 62 | 72 | 55 | 73 | 23 | | 17389598 | - | - |
| Dlc1 (Q63744) | Rattus norvegicus | S807 | NTLKRENSSPRVMQR | - | 0.5055 | TP | certain | experimental | 55 | 30 | 72 | 148 | 9 | | 21087603 | - | - |
| SNAI1 (O95863) | Homo sapiens | S11 | SFLVRKPSDPNRKPN | 0.356 | 0.6948 | TP | certain | experimental | 66 | 72 | 73 | 8 | 55 | | 20940406 | - | - |
| MARK2 (Q7KZI7) | Homo sapiens | S400 | HKVQRSVSANPKQRR | 0.995 | 0.8279 | TP | certain | experimental | 73 | 9 | 55 | 107 | 72 | | 19011111 | - | - |
| VASP (P50552) | Homo sapiens | S157 | EHIERRVSNAGGPPA | 0.401 | 0.9473 | TP | certain | experimental | 72 | 147 | 9 | 55 | 73 | 16 | | 23846685 | - | - |
| VASP (P50552) | Homo sapiens | S322 | TTLPRMKSSSSVTTS | 0.364 | 0.8596 | TP | certain | experimental | 72 | 9 | 55 | 73 | 148 | 16 | | 23846685 | - | - |
| PAK4 (O96013) | Homo sapiens | S99 | MSVTRSNSLRRDSPP | 0.968 | 0.892 | TP | certain | experimental | 72 | 55 | 9 | 73 | 39 | | 23841590 | - | - |
| PAK4 (O96013) | Homo sapiens | S474 | KEVPRRKSLVGTPYW | 0.786 | 0.2951 | TP | certain | experimental | 72 | 55 | 73 | 148 | | 21832093 | - | - |
| Evl (P70429) | Mus musculus | S343 | SLLSRTPSVAKSPEA | - | 0.8421 | TP | certain | experimental | 72 | 73 | 9 | 55 | 40 | 23 | | 19000756 | - | - |
| RIN1 (Q13671) | Homo sapiens | S292 | QLLRRESSVGYRVPA | 0.737 | 0.5456 | TP | certain | experimental | 55 | 62 | 73 | 30 | 72 | 148 | 39 | | 21209314 | - | - |
| RIN1 (Q13671) | Homo sapiens | S351 | RPLLRSMSAAFCSLL | 0.732 | 0.391 | TP | certain | experimental | 72 | 9 | 30 | 55 | | 21209314 | - | - |
| PI4KB (Q9UBF8) | Homo sapiens | S294 | SNLKRTASNPKVENE | 0.332 | 0.7459 | TP | certain | experimental | 72 | 9 | 55 | 73 | 74 | | 16100512 | - | - |
| Kidins220 (Q9EQG6) | Rattus norvegicus | S918 | RTITRQMSFDLTKLL | - | 0.2752 | TP | certain | experimental | 23 | 72 | 73 | 9 | 55 | 131 | | 10998417 | - | - |
| SSH1 (Q8WYL5) | Homo sapiens | S978 | SPLKRSHSLAKLGSL | 0.802 | 0.4369 | TP | certain | experimental | 72 | 73 | 55 | 9 | 143 | | 19329994 | - | - |
| HSPB1 (P04792) | Homo sapiens | S82 | RALSRQLSSGVSEIR | 0.622 | 0.374 | TP | certain | experimental | 55 | 72 | 9 | 73 | | 15728188 | - | - |
| EGFR (P00533) | Homo sapiens | T678 | RHIVRKRTLRRLLQE | 0.415 | 0.3182 | TP | certain | experimental | 72 | 9 | | 10523301 | - | - |
| EGFR (P00533) | Homo sapiens | T693 | RELVEPLTPSGEAPN | 0.275 | 0.4476 | TP | certain | experimental | 72 | 9 | | 10523301 | - | - |
| CTNNB1 (P35222) | Homo sapiens | T112 | EGMQIPSTQFDAAHP | 0.535 | 0.6375 | TP | certain | experimental | 66 | 72 | 135 | 62 | 23 | 73 | 147 | | 19141652 | - | - |
| CTNNB1 (P35222) | Homo sapiens | T120 | QFDAAHPTNVQRLAE | 0.721 | 0.7755 | TP | certain | experimental | 66 | 72 | 135 | 62 | 23 | 73 | 147 | | 19141652 | - | - |
| PRKD1 (Q15139) | Homo sapiens | S205 | GVRRRRLSNVSLTGV | 0.511 | 0.2328 | - | - | - | - | - | Non high throughput (10867018) | - |
| Prkd1 (Q62101) | Mus musculus | S203 | GVRRRRLSNVSLTGL | - | 0.2258 | - | - | - | - | - | Non high throughput (10867018) | - |
| PPP1R14A (Q96A00) | Homo sapiens | T38 | QKRHARVTVKYDRRE | 0.852 | 0.4979 | - | - | - | - | - | Non high throughput (15003508) |
|
| Prkd1 (Q62101) | Mus musculus | S916 | KALSERVSIL_____ | - | 0.2258 | - | - | - | - | - | Non high throughput (10473617, 10867018) | - |
| PRKD1 (Q15139) | Homo sapiens | S910 | KALGERVSIL_____ | 0.500 | 0.2849 | - | - | - | - | - | Non high throughput (10473617, 14607845, 16563698) |
|
| Prkd1 (Q62101) | Mus musculus | S748 | GEKSFRRSVVGTPAY | - | 0.1323 | - | - | - | - | - | Non high throughput (10867018, 12407104) | - |
| Prkd1 (Q62101) | Mus musculus | S744 | ARIIGEKSFRRSVVG | - | 0.0817 | - | - | - | - | - | Non high throughput (10867018, 2407104) | - |
| HDAC7 (Q8WUI4) | Homo sapiens | S155 | FPLRKTVSEPNLKLR | 0.972 | 0.6227 | - | - | - | - | - | Non high throughput (15623513, 18509061, 18617643) |
|
| TLR4 (O00206) | Homo sapiens | S790 | VELYRLLSRNTYLEW | 0.047 | 0.0543 | - | - | - | - | - | - |
|
| TLR5 (O60602) | Homo sapiens | S805 | YQLMKHQSIRGFVQK | 0.044 | 0.1007 | - | - | - | - | - | - |
|
| MBP (P02686-5) | Homo sapiens | S162 | FKLGGRDSRSGSPMA | 0.116 | 0.7036 | - | - | - | - | - | - |
|
| JUN (P05412) | Homo sapiens | S63 | KNSDLLTSPDVGLLK | 0.767 | 0.3426 | - | - | - | - | - | - |
|
| CACNA1C (P15381) | Oryctolagus cuniculus | S1928 | ASLGRRASFHLECLK | - | 0.3392 | - | - | - | - | - | - |
|
| OSBP (P22059) | Homo sapiens | S240 | TALQRSLSELESLKL | 0.351 | 0.4652 | - | - | - | - | - | - |
|
| CFL1 (P23528) | Homo sapiens | S3 | _____MASGVAVSDG | 0.971 | 0.1611 | - | - | - | - | - | - |
|
| Tnni3 (P23693) | Rattus norvegicus | S23 | PAPVRRRSSANYRAY | - | 0.6755 | - | - | - | - | - | - |
|
| Tnni3 (P23693) | Rattus norvegicus | S24 | APVRRRSSANYRAYA | - | 0.708 | - | - | - | - | - | - |
|
| Pik3r1 (P26450) | Mus musculus | S652 | TFLVRESSKQGCYAC | - | 0.2002 | - | - | - | - | - | - |
|
| AKT1 (P31749) | Homo sapiens | S473 | RPHFPQFSYSASGTA | 0.952 | 0.5342 | - | - | - | - | - | - |
|
| Syk (P48025) | Mus musculus | S291 | ISRIKSYSFPKPGHK | - | 0.4292 | - | - | - | - | - | - |
|
| Tnni3 (P48787) | Mus musculus | S23 | PAPVRRRSSANYRAY | - | 0.6906 | - | - | - | - | - | - |
|
| Tnni3 (P48787) | Mus musculus | S24 | APVRRRSSANYRAYA | - | 0.7163 | - | - | - | - | - | - |
|
| CACNA1C (Q13936) | Homo sapiens | S1981 | ASLGRRASFHLECLK | 0.889 | 0.384 | - | - | - | - | - | - |
|
| CTTN (Q14247) | Homo sapiens | S348 | EAVTSKTSNIRANFE | 0.455 | 0.5382 | - | - | - | - | - | - |
|
| MYBPC3 (Q14896) | Homo sapiens | S304 | SLLKKRDSFRTPRDS | 0.236 | 0.3704 | - | - | - | - | - | - |
|
| PRKD1 (Q15139) | Homo sapiens | S742 | GEKSFRRSVVGTPAY | 0.714 | 0.1323 | - | - | - | - | - | - |
|
| Ahcyl1 (Q80SW1) | Mus musculus | S68 | RSLSRSISQSSTDSY | - | 0.5854 | - | - | - | - | - | - |
|
| HDAC7 (Q8WUI4) | Homo sapiens | S358 | WPLSRTRSEPLPPSA | 0.984 | 0.8013 | - | - | - | - | - | - |
|
| HDAC7 (Q8WUI4) | Homo sapiens | S486 | RPLSRAQSSPAAPAS | 0.983 | 0.805 | - | - | - | - | - | - |
|
| SSH1 (Q8WYL5) | Homo sapiens | S402 | MGVSRSASTVIAYAM | 0.666 | 0.1611 | - | - | - | - | - | - |
|
| SSH1 (Q8WYL5) | Homo sapiens | S937 | SNLTRSSSSDSIHSV | 0.839 | 0.6948 | - | - | - | - | - | - |
|
| MAP4K1 (Q92918) | Homo sapiens | S171 | ATLARRLSFIGTPYW | 0.778 | 0.0454 | - | - | - | - | - | - |
|
| BAD (Q92934) | Homo sapiens | S118 | GRELRRMSDEFVDSF | 0.787 | 0.5758 | - | - | - | - | - | - |
|
| BAD (Q92934) | Homo sapiens | S75 | EIRSRHSSYPAGTED | 0.509 | 0.8198 | - | - | - | - | - | - |
|
| BAD (Q92934) | Homo sapiens | S99 | PFRGRSRSAPPNLWA | 0.900 | 0.7982 | - | - | - | - | - | - |
|
| DLC1 (Q96QB1) | Homo sapiens | S1244 | NTLKRENSSPRVMQR | 0.595 | 0.4979 | - | - | - | - | - | - |
|
| SPHK2 (Q9NRA0) | Homo sapiens | S419 | AHSPLHRSVSDLPLP | 0.402 | 0.6712 | - | - | - | - | - | - |
|
| SPHK2 (Q9NRA0) | Homo sapiens | S421 | SPLHRSVSDLPLPLP | 0.657 | 0.5992 | - | - | - | - | - | - |
|
| PI4KB (Q9UBF8-2) | Homo sapiens | S294 | SNLKRTASNPKVENE | 0.332 | 0.7547 | - | - | - | - | - | - |
|
| HDAC5 (Q9UQL6) | Homo sapiens | S259 | FPLRKTASEPNLKVR | 0.835 | 0.6712 | - | - | - | - | - | - |
|
| HDAC5 (Q9UQL6) | Homo sapiens | S498 | RPLSRTQSSPLPQSP | 0.940 | 0.9126 | - | - | - | - | - | - |
|
| PTRH2 (Q9Y3E5) | Homo sapiens | S5 | ___MPSKSLVMEYLA | 0.304 | 0.2645 | - | - | - | - | - | - |
|
| PTRH2 (Q9Y3E5) | Homo sapiens | S87 | GKVAAQCSHAAVSAY | 0.357 | 0.1702 | - | - | - | - | - | - |
|
| AKT1 (P31749) | Homo sapiens | T308 | KDGATMKTFCGTPEY | 0.710 | 0.2645 | - | - | - | - | - | - |
|
| PIP4K2A (P48426) | Homo sapiens | T376 | KAAHAAKTVKHGAGA | 0.780 | 0.5176 | - | - | - | - | - | - |
|
| TCAP (O15273) | Homo sapiens | S157 | GALRRSLSRSMSQEA | 0.250 | 0.6806 | - | - | - | - | - | - |
|
| Prkaa1 (Q5EG47) | Mus musculus | S494 | GTATPQRSGSISNYR | - | 0.5533 | - | - | - | - | - | - |
|
| DNM1L (O00429) | Homo sapiens | S637 | VPVARKLSAREQRDC | 0.300 | 0.4119 | - | - | - | - | - | - |
|
| Prkaa2 (Q8BRK8) | Mus musculus | S491 | STPQRSCSAAGLHRA | - | 0.5419 | - | - | - | - | - | - |
|
| Cdh2 (Q9Z1Y3) | Rattus norvegicus | S869 | GSGSTAGSLSSLNSS | - | 0.7331 | - | - | - | - | - | - |
|
| RABEP1 (Q15276) | Homo sapiens | S407 | DGLRRAQSTDSLGTS | 0.942 | 0.5342 | - | - | - | - | - | - |
|
| REM1 (O75628) | Homo sapiens | S18 | TPLHRRASTPLPLSP | 0.999 | 0.8681 | - | - | - | - | - | - |
|
| Plekhg5 (Q66T02) | Mus musculus | S92 | SVLARNVSTRSCPPR | - | 0.6991 | - | - | - | - | - | - |
|
| Plekhg5 (Q66T02) | Mus musculus | S806 | PTILRKSSNSLDSEH | - | 0.5665 | - | - | - | - | - | - |
|
| OSBP (P16258) | Oryctolagus cuniculus | S242 | TALQRSLSELESLKL | - | 0.4582 | - | - | - | - | - | - |
|
| Cdh2 (Q9Z1Y3) | Rattus norvegicus | S871 | GSTAGSLSSLNSSSS | - | 0.8125 | - | - | - | - | - | - |
|
| Drd1 (P18901) | Rattus norvegicus | S421 | EKLSPALSVILDYDT | - | 0.2064 | - | - | - | - | - | - |
|
| Mybpc3 (O70468) | Mus musculus | S302 | SLLKKRDSFRRDSKL | - | 0.384 | - | - | - | - | - | - |
|
| RTKN (Q9BST9-2) | Homo sapiens | S435 | QALAKQGSLYHEMAI | 0.149 | 0.4979 | - | - | - | - | - | - |
|
| ARFIP1 (P53367) | Homo sapiens | S132 | LELVRKWSLNTYKCT | 0.753 | 0.1914 | - | - | - | - | - | - |
|
| Cdh2 (Q9Z1Y3) | Rattus norvegicus | S872 | STAGSLSSLNSSSSG | - | 0.8125 | - | - | - | - | - | - |
|
| ARFIP1 (P53367-2) | Homo sapiens | S100 | LELVRKWSLNTYKCT | 0.588 | 0.2034 | - | - | - | - | - | - |
|
| Nlrp3 (Q8R4B8) | Mus musculus | S291 | CKILRKPSRILFLMD | - | 0.0433 | - | - | - | - | - | - |
|
| HSPB6 (O14558) | Homo sapiens | S16 | PSWLRRASAPLPGLS | 0.206 | 0.4409 | - | - | - | - | - | - |
|
| TCAP (O15273) | Homo sapiens | S161 | RSLSRSMSQEAQRG_ | 0.844 | 0.5665 | - | - | - | - | - | - |
|
| MAP3K5 (Q99683) | Homo sapiens | T838 | GINPCTETFTGTLQY | 0.596 | 0.2328 | - | - | - | - | - | - |
|
| ITGB4 (P16144-2) | Homo sapiens | T1736 | EFVSRTLTTSGTLST | 0.859 | 0.4864 | - | - | - | - | - | - |
|
| PLEKHG5 (O94827-7) | Homo sapiens | S92 | SVLARNVSTRSCPPR | 0.764 | 0.7289 | - | - | - | - | - | - | - |
| CACNA1C (Q13936-9) | Homo sapiens | S1898 | LLRKANPSRCHSRES | 0.720 | 0.4801 | - | - | - | - | - | - | - |
| AHCYL1 (O43865) | Homo sapiens | S68 | RSLSRSISQSSTDSY | 0.608 | 0.5854 | - | - | - | - | - | - | - |
| TNNI3 (P19429) | Homo sapiens | S23 | PAPIRRRSSNYRAYA | 0.104 | 0.6906 | - | - | - | - | - | - | - |
| TNNI3 (P19429) | Homo sapiens | S24 | APIRRRSSNYRAYAT | 0.016 | 0.7163 | - | - | - | - | - | - | - |
| PIK3R1 (P27986) | Homo sapiens | S652 | TFLVRESSKQGCYAC | 0.443 | 0.2002 | - | - | - | - | - | - | - |
| SYK (P43405) | Homo sapiens | S291 | WSAGGIISRIKSYSF | 0.497 | 0.3053 | - | - | - | - | - | - | - |
| PRKD1 (Q15139) | Homo sapiens | S738 | ARIIGEKSFRRSVVG | 0.590 | 0.0817 | - | - | - | - | - | - | - |
| TRPV1 (Q8NER1) | Homo sapiens | S117 | LRLYDRRSIFEAVAQ | 0.045 | 0.3019 | - | - | - | - | - | - | - |
| HDAC7 (Q8WUI4-5) | Homo sapiens | S194 | FPLRKTVSEPNLKLR | 0.194 | 0.6227 | - | - | - | - | - | - | - |
| HDAC7 (Q8WUI4-5) | Homo sapiens | S220 | NPLLRKESAPPSLRR | 0.385 | 0.8343 | - | - | - | - | - | - | - |
| HDAC7 (Q8WUI4-5) | Homo sapiens | S397 | WPLSRTRSEPLPPSA | 0.646 | 0.8013 | - | - | - | - | - | - | - |
| RTKN (Q9BST9) | Homo sapiens | S448 | QALAKQGSLYHEMAI | 0.952 | 0.4979 | - | - | - | - | - | - | - |
| HDAC5 (Q9UQL6-3) | Homo sapiens | S260 | FPLRKTASEPNLKVR | 0.838 | 0.6712 | - | - | - | - | - | - | - |
| HDAC5 (Q9UQL6-3) | Homo sapiens | S499 | RPLSRTQSSPLPQSP | 0.120 | 0.9126 | - | - | - | - | - | - | - |
| SYK (A0A024R244) | Homo sapiens | S297 | ISRIKSYSFPKPGHR | - | 0.4186 | - | - | - | - | - | - | - |
| PRKD1 (F8WBA3) | Homo sapiens | S205 | GVRRRRLSNVSLTGV | - | 0.2399 | - | - | - | - | - | - | - |
| PRKD1 (F8WBA3) | Homo sapiens | S746 | ARIIGEKSFRRSVVG | - | 0.0817 | - | - | - | - | - | - | - |
| PRKD1 (F8WBA3) | Homo sapiens | S750 | GEKSFRRSVVGTPAY | - | 0.1323 | - | - | - | - | - | - | - |
| PRKD1 (F8WBA3) | Homo sapiens | S918 | KALGERVSIL_____ | - | 0.2849 | - | - | - | - | - | - | - |
| AHCYL1 (A0A024R0A8) | Homo sapiens | S68 | RSLSRSISQSSTDSY | - | 0.5854 | - | - | - | - | - | - | - |
| KIDINS220 (Q9ULH0) | Homo sapiens | S918 | RTITRQMSFDLTKLL | 0.187 | 0.268 | - | - | - | - | - | - | - |
| DLC1 (Q96QB1) | Homo sapiens | S733 | VPMVRKRSVSNSTQT | 0.731 | 0.662 | - | - | - | - | - | - | - |
| EVL (Q9UI08) | Homo sapiens | S345 | SILSRTPSVAKSPEA | 0.770 | 0.8235 | - | - | - | - | - | - | - |
| PLCG1 variant protein (Q4LE43) | Homo sapiens | S1370 | HGRAREGSFESRYQQ | - | 0.384 | - | - | - | - | - | - | - |
| PRKAA1 (Q13131) | Homo sapiens | S494 | GTATPQRSGSVSNYR | 0.112 | 0.5758 | - | - | - | - | - | - | - |
| (B2RA25) | Homo sapiens | S491 | STPQRSCSAAGLHRP | - | 0.6531 | - | - | - | - | - | - | - |
| DRD1 (P21728) | Homo sapiens | S421 | EKLSPALSVILDYDT | 0.623 | 0.2164 | - | - | - | - | - | - | - |
| MYBPC3 (A8MXZ9) | Homo sapiens | S304 | SLLKKSSSFRTPRDS | - | 0.3631 | - | - | - | - | - | - | - |
| CACNA1C (Q13936) | Homo sapiens | T1928 | EVKMNHDTEACSEPS | 0.098 | 0.5456 | - | - | - | - | - | - | - |
Help Instructions
- Hovering on the "*" in "Substrate" column will display the text if there is a note by the annotator for the given site.
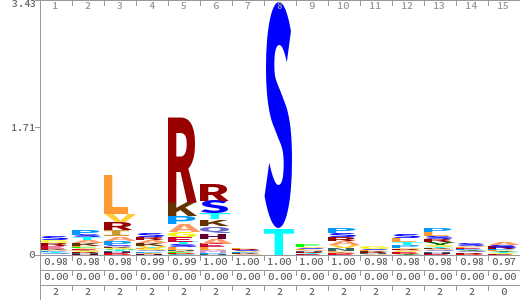
- The column having "Sequence" motif represents phosphosite and its neighbouring 7 residues (upstream and downstream).
- The score in "Conservation" column varies from 0 to 1; Higher the score more conserved is the site.
- The score in "Disorder" column varies from 0 to 1; Score higher than 0.4 suggests disorder.
- In the "Curator Assessment" column, TP refers to True Positive; FP refers to False Positive; and U refers to unknown if the annoatator cannot decide the status based on available information.
- Hovering on the number in the "Evidence Logic" column will show the name of method used to characterize/infer the phosphosite (for more details plase follow the link)