Help Instruction
- Double clicking on the substrate will redirect to the uniprot entry.
- Clicking in the block having the substrate will zoom into its phophosites
- Clicking the center of the circle will lead to zooming out.
- Clicking on the phosphosite will render the phosphosite in its 3D structure (if structure information/mapping is available).
- The color of every substrate has ben kept different in the Sunburst diagram for visual clarity.
- Clicking in the top right corner on the camara icon will download the image.
| Substrate | Organism | PSite | Sequence(+/-7) | Conservation | Disorder | Curator Assessment | Reliability | Evidence Class | Evidence Logic | PubMed | Phospho-ELM | PhosphoSite-Plus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PPP1R3A (Q16821) | Homo sapiens | S48 | PSRRGSDSSEDIYLD | 0.774 | 0.7331 | - | - | - | - | - | Non high throughput (2152882) | - |
| ESR2 (Q92731) | Homo sapiens | S165 | GYHYGVWSCEGCKAF | 0.777 | 0.0281 | - | - | - | - | - | Non high throughput (9528769) | - |
| CEBPB (P17676) | Homo sapiens | T266 | VKSKAKKTVDKHSDE | 0.154 | 0.6755 | - | - | - | - | - | Non high throughput (10635333) | - |
| BAD (Q92934) | Homo sapiens | S118 | GRELRRMSDEFVDSF | 0.787 | 0.5758 | - | - | - | - | - | Non high throughput (10837486) | - |
| BAD (Q92934) | Homo sapiens | S75 | EIRSRHSSYPAGTED | 0.509 | 0.8198 | - | - | - | - | - | Non high throughput (10837486) |
|
| ETV1 (P50549) | Homo sapiens | S191 | HRFRRQLSEPCNSFP | 0.258 | 0.6043 | - | - | - | - | - | Non high throughput (12213813) | - |
| ETV1 (P50549) | Homo sapiens | S216 | PMYQRQMSEPNIPFP | 0.364 | 0.8313 | - | - | - | - | - | Non high throughput (12213813) | - |
| ETV1 (P50549) | Homo sapiens | S334 | PTYQRRGSLQLWQFL | 0.576 | 0.1702 | - | - | - | - | - | Non high throughput (12213813) | - |
| NFKBIB (Q15653) | Homo sapiens | S19 | DADEWCDSGLGSLGP | 0.929 | 0.3529 | - | - | - | - | - | Non high throughput (16822942) | - |
| NFKBIB (Q15653) | Homo sapiens | S23 | WCDSGLGSLGPDAAA | 0.928 | 0.4119 | - | - | - | - | - | Non high throughput (16822942) | - |
| IRS1 (P35568) | Homo sapiens | S1101 | GCRRRHSSETFSSTP | 0.923 | 0.8085 | - | - | - | - | - | Non high throughput (17709744) | - |
| EEF2K (O00418) | Homo sapiens | S366 | SPQVRTLSGSRPPLL | 0.742 | 0.6227 | - | - | - | - | - | - |
|
| MITF (O75030-9) | Homo sapiens | S409 | KTSSRRSSMSMEETE | 0.697 | 0.7289 | - | - | - | - | - | - |
|
| FOS (P01100) | Homo sapiens | S362 | AAAHRKGSSSNEPSS | 0.803 | 0.5758 | - | - | - | - | - | - |
|
| ESR1 (P03372) | Homo sapiens | S167 | GGRERLASTNDKGSM | 0.551 | 0.7459 | - | - | - | - | - | - |
|
| Nr4a1 (P12813) | Mus musculus | S354 | GRRGRLPSKPKQPPD | - | 0.7755 | - | - | - | - | - | - |
|
| ITGB4 (P16144) | Homo sapiens | S1364 | PSGSQRPSVSDDTGC | 0.798 | 0.6948 | - | - | - | - | - | - |
|
| CREB1 (P16220) | Homo sapiens | S133 | EILSRRPSYRKILND | - | 0.4979 | - | - | - | - | - | - |
|
| SLC9A1 (P19634) | Homo sapiens | S703 | MSRARIGSDPLAYEP | 0.834 | 0.7799 | - | - | - | - | - | - |
|
| FLNA (P21333) | Homo sapiens | S2152 | TRRRRAPSVANVGSH | 0.180 | 0.494 | - | - | - | - | - | - |
|
| EIF4B (P23588) | Homo sapiens | S406 | RPRERHPSWRSEETQ | 0.581 | 0.8462 | - | - | - | - | - | - | - |
| EIF4B (P23588) | Homo sapiens | S422 | RERSRTGSESSQTGT | 0.591 | 0.9457 | - | - | - | - | - | - |
|
| NFKBIA (P25963) | Homo sapiens | S32 | LLDDRHDSGLDSMKD | 0.999 | 0.6089 | - | - | - | - | - | - |
|
| L1CAM (P32004) | Homo sapiens | S1152 | RSKGGKYSVKDKEDT | 0.246 | 0.4087 | - | - | - | - | - | - | - |
| TSC2 (P49815) | Homo sapiens | S1798 | GQRKRLISSVEDFTE | 0.406 | 0.247 | - | - | - | - | - | - |
|
| Tsc2 (P49816) | Rattus norvegicus | S939 | KFRARSTSLNERPKS | - | 0.4369 | - | - | - | - | - | - |
|
| GSK3B (P49841) | Homo sapiens | S9 | SGRPRTTSFAESCKP | 0.500 | 0.5951 | - | - | - | - | - | - |
|
| DAPK1 (P53355) | Homo sapiens | S289 | QALSRKASAVNMEKF | - | 0.3356 | - | - | - | - | - | - |
|
| RPS6 (P62753) | Homo sapiens | S235 | IAKRRRLSSLRASTS | 0.272 | 0.6991 | - | - | - | - | - | - |
|
| RPS6 (P62753) | Homo sapiens | S236 | AKRRRLSSLRASTSK | 0.934 | 0.6906 | - | - | - | - | - | - |
|
| YBX1 (P67809) | Homo sapiens | S102 | NPRKYLRSVGDGETV | 0.503 | 0.6712 | - | - | - | - | - | - |
|
| CCT2 (P78371) | Homo sapiens | S260 | GSRVRVDSTAKVAEI | 0.394 | 0.1942 | - | - | - | - | - | - |
|
| MDM2 (Q00987) | Homo sapiens | S166 | SSRRRAISETEENSD | 0.815 | 0.7718 | - | - | - | - | - | - | - |
| MDM2 (Q00987) | Homo sapiens | S186 | RQRKRHKSDSISLSF | 0.958 | 0.6043 | - | - | - | - | - | - | - |
| MXD1 (Q05195) | Homo sapiens | S145 | IERIRMDSIGSTVSS | 0.924 | 0.4901 | - | - | - | - | - | - |
|
| Mitf (Q08874-8) | Mus musculus | S409 | KTSSRRSSMSAEETE | - | 0.712 | - | - | - | - | - | - |
|
| RPS6KA1 (Q15418) | Homo sapiens | S221 | DHEKKAYSFCGTVEY | 0.758 | 0.1611 | - | - | - | - | - | - |
|
| Bad (Q61337) | Mus musculus | S112 | ETRSRHSSYPAGTEE | - | 0.8828 | - | - | - | - | - | - |
|
| Bad (Q61337) | Mus musculus | S136 | PFRGRSRSAPPNLWA | - | 0.6661 | - | - | - | - | - | - |
|
| Bad (Q61337) | Mus musculus | S155 | GRELRRMSDEFEGSF | - | 0.5456 | - | - | - | - | - | - |
|
| Kcnd3 (Q62897) | Rattus norvegicus | S535 | YPSTRSPSLSSHSGL | - | 0.7415 | - | - | - | - | - | - |
|
| Kcnd3 (Q62897) | Rattus norvegicus | S569 | LPATRLRSMQELSTI | - | 0.5758 | - | - | - | - | - | - |
|
| Rps6ka1 (Q63531) | Rattus norvegicus | S221 | DHEKKAYSFCGTVEY | - | 0.1583 | - | - | - | - | - | - |
|
| Rps6ka1 (Q63531) | Rattus norvegicus | S380 | HQLFRGFSFVATGLM | - | 0.1942 | - | - | - | - | - | - |
|
| Rps6ka1 (Q63531) | Rattus norvegicus | S732 | RRVRKLPSTTL____ | - | 0.4725 | - | - | - | - | - | - |
|
| Dlc1 (Q63744) | Rattus norvegicus | S330 | VTRTRSLSTCNKRVG | - | 0.5055 | - | - | - | - | - | - |
|
| Eif4b (Q8BGD9) | Mus musculus | S422 | RERSRTGSESSQTGA | - | 0.9473 | - | - | - | - | - | - |
|
| RPTOR (Q8N122) | Homo sapiens | S719 | PCTPRLRSVSSYGNI | 0.155 | 0.3774 | - | - | - | - | - | - |
|
| RPTOR (Q8N122) | Homo sapiens | S721 | TPRLRSVSSYGNIRA | 0.834 | 0.3392 | - | - | - | - | - | - |
|
| RPTOR (Q8N122) | Homo sapiens | S722 | PRLRSVSSYGNIRAV | 0.794 | 0.3149 | - | - | - | - | - | - |
|
| DEPTOR (Q8TB45) | Homo sapiens | S286 | SSMSSCGSSGYFSSS | 0.184 | 0.3392 | - | - | - | - | - | - |
|
| DEPTOR (Q8TB45) | Homo sapiens | S287 | SMSSCGSSGYFSSSP | 0.801 | 0.3872 | - | - | - | - | - | - |
|
| DEPTOR (Q8TB45) | Homo sapiens | S291 | CGSSGYFSSSPTLSS | 0.165 | 0.4186 | - | - | - | - | - | - |
|
| CIC (Q96RK0) | Homo sapiens | S173 | PGKRRTQSLSALPKE | 0.360 | 0.8872 | - | - | - | - | - | - |
|
| RANBP3 (Q9H6Z4) | Homo sapiens | S126 | VKRERTSSLTQFPPS | 0.804 | 0.8741 | - | - | - | - | - | - |
|
| Krt17 (Q9QWL7) | Mus musculus | S44 | AGSCRLGSASGLGSA | - | 0.247 | - | - | - | - | - | - |
|
| METTL1 (Q9UBP6) | Homo sapiens | S27 | YYRQRAHSNPMADHT | 0.762 | 0.4652 | - | - | - | - | - | - |
|
| NDRG2 (Q9UN36) | Homo sapiens | S332 | LSRSRTASLTSAASV | 0.542 | 0.5456 | - | - | - | - | - | - |
|
| NDRG2 (Q9UN36) | Homo sapiens | S350 | RSRSRTLSQSSESGT | 0.882 | 0.7755 | - | - | - | - | - | - |
|
| CARHSP1 (Q9Y2V2) | Homo sapiens | S52 | TRRTRTFSATVRASQ | 0.908 | 0.6806 | - | - | - | - | - | - |
|
| Nos1 (Q9Z0J4) | Mus musculus | S847 | SYKVRFNSVSSYSDS | - | 0.3356 | - | - | - | - | - | - | - |
| Cebpb (P21272) | Rattus norvegicus | S105 | AKPSKKPSDYGYVSL | - | 0.3872 | - | - | - | - | - | - | - |
| CDKN1B (P46527) | Homo sapiens | T198 | PGLRRRQT_______ | 0.507 | 0.9488 | - | - | - | - | - | - | - |
| Tsc2 (P49816) | Rattus norvegicus | T1466 | GLRPRGYTISDSAPS | - | 0.7415 | - | - | - | - | - | - |
|
| VASP (P50552) | Homo sapiens | T278 | LARRRKATQVGEKTP | 0.261 | 0.8828 | - | - | - | - | - | - |
|
| GMFB (P60983) | Homo sapiens | T27 | KFRFRKETNNAAIIM | 0.307 | 0.1528 | - | - | - | - | - | - |
|
| Cebpb (P28033) | Mus musculus | T217 | AKAKAKKTVDKLSDE | - | 0.5456 | - | - | - | - | - | - | - |
| UBR5 (O95071) | Homo sapiens | S2483 | ENRKRHGSSRSVVDM | 0.274 | 0.5493 | - | - | - | - | - | - |
|
| Mapk7 (Q9WVS8) | Mus musculus | S496 | SRLRDGPSAPLEAPE | - | 0.8828 | - | - | - | - | - | - |
|
| MAPK7 (Q13164) | Homo sapiens | S496 | SRLRDGPSAPLEAPE | 0.093 | 0.8894 | - | - | - | - | - | - |
|
| WWC1 (Q8IX03) | Homo sapiens | S947 | CRLNRSDSDSSTLSK | 0.881 | 0.6322 | - | - | - | - | - | - |
|
| Gab2 (Q9Z1S8) | Mus musculus | S160 | LLRERKSSAPSHSSQ | - | 0.6906 | - | - | - | - | - | - |
|
| CDC25C (P30307) | Homo sapiens | S247 | KVKKKYFSGQGKLRK | 0.394 | 0.2364 | - | - | - | - | - | - |
|
| CHEK1 (O14757) | Homo sapiens | S280 | AKRPRVTSGGVSESP | 0.246 | 0.6227 | - | - | - | - | - | - |
|
| PDCD4 (Q53EL6) | Homo sapiens | S457 | RGRKRFVSEGDGGRL | 0.922 | 0.4507 | - | - | - | - | - | - |
|
| SOS1 (Q07889) | Homo sapiens | S1134 | PHGPRSASVSSISLT | 0.446 | 0.7547 | - | - | - | - | - | - |
|
| LCP1 (P13796) | Homo sapiens | S5 | ___MARGSVSDEEMM | 0.166 | 0.4476 | - | - | - | - | - | - |
|
| Gab2 (Q9Z1S8) | Mus musculus | S620 | PSPHRKPSTSSVTSD | - | 0.8942 | - | - | - | - | - | - |
|
| APAF1 (O14727) | Homo sapiens | S357 | FKRIRKSSSYDYEAL | 0.525 | 0.2292 | - | - | - | - | - | - |
|
| SOS1 (Q07889) | Homo sapiens | S1161 | PPRRRPESAPAESSP | 0.496 | 0.9308 | - | - | - | - | - | - |
|
| UBR5 (O95071) | Homo sapiens | S1227 | DCKLKRTSPTAYCDC | 0.557 | 0.0646 | - | - | - | - | - | - |
|
| Senp2 (Q91ZX6) | Mus musculus | S38 | RSSSTLFSTAVDTDE | - | 0.4441 | - | - | - | - | - | - |
|
| OSTF1 (Q92882) | Homo sapiens | S202 | TDAVRTLSNAEDYLD | 0.325 | 0.4766 | - | - | - | - | - | - |
|
| CDC25A (P30304) | Homo sapiens | S295 | TKRRKSMSGASPKES | 0.296 | 0.8792 | - | - | - | - | - | - |
|
| PDCD4 (Q53EL6) | Homo sapiens | S76 | RDSGRGDSVSDSGSD | 0.546 | 0.7755 | - | - | - | - | - | - |
|
| EPHA2 (P29317) | Homo sapiens | S897 | RVSIRLPSTSGSEGV | 0.425 | 0.3356 | - | - | - | - | - | - |
|
| CDC25A (P30304) | Homo sapiens | S293 | GSTKRRKSMSGASPK | 0.631 | 0.8713 | - | - | - | - | - | - |
|
| Gab2 (Q9Z1S8) | Mus musculus | S211 | TENARSASFSQGTRQ | - | 0.7881 | - | - | - | - | - | - |
|
| APAF1 (O14727) | Homo sapiens | S268 | LLTTRDKSVTDSVMG | 0.653 | 0.2034 | - | - | - | - | - | - |
|
| CDC25B (P30305) | Homo sapiens | S353 | VQNKRRRSVTPPEEQ | 0.838 | 0.8713 | - | - | - | - | - | - |
|
| CDC25B (P30305) | Homo sapiens | T355 | NKRRRSVTPPEEQQE | 0.138 | 0.8765 | - | - | - | - | - | - |
|
| UBR5 (O95071) | Homo sapiens | T637 | PYKRRRSTPAPKEEE | 0.124 | 0.7595 | - | - | - | - | - | - |
|
| Senp2 (Q91ZX6) | Mus musculus | T368 | TEDLFEFTEDMEKEI | - | 0.4292 | - | - | - | - | - | - |
|
| Senp2 (Q91ZX6) | Mus musculus | T35 | KRRRSSSTLFSTAVD | - | 0.3494 | - | - | - | - | - | - |
|
| SLC9A3R1 (O14745) | Homo sapiens | T156 | ELRPRLCTMKKGPSG | 0.071 | 0.5296 | - | - | - | - | - | - |
|
| CDC34 (P49427) | Homo sapiens | T162 | KGKDREYTDIIRKQV | 0.018 | 0.4725 | - | - | - | - | - | - |
|
| SENP2 (Q9HC62) | Homo sapiens | T369 | TDDLLELTEDMEKEI | 0.314 | 0.4766 | - | - | - | - | - | - |
|
| WWC1 (Q8IX03) | Homo sapiens | T929 | STIIRSKTFSPGPQS | 0.849 | 0.5493 | - | - | - | - | - | - |
|
Help Instructions
- Hovering on the "*" in "Substrate" column will display the text if there is a note by the annotator for the given site.
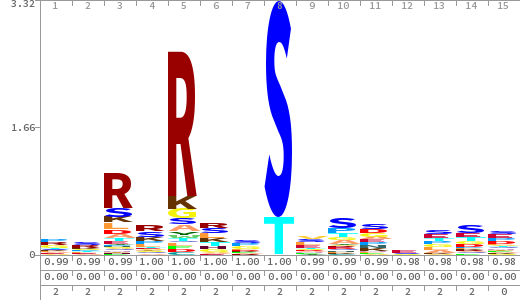
- The column having "Sequence" motif represents phosphosite and its neighbouring 7 residues (upstream and downstream).
- The score in "Conservation" column varies from 0 to 1; Higher the score more conserved is the site.
- The score in "Disorder" column varies from 0 to 1; Score higher than 0.4 suggests disorder.
- In the "Curator Assessment" column, TP refers to True Positive; FP refers to False Positive; and U refers to unknown if the annoatator cannot decide the status based on available information.
- Hovering on the number in the "Evidence Logic" column will show the name of method used to characterize/infer the phosphosite (for more details plase follow the link)