Help Instruction
- Double clicking on the substrate will redirect to the uniprot entry.
- Clicking in the block having the substrate will zoom into its phophosites
- Clicking the center of the circle will lead to zooming out.
- Clicking on the phosphosite will render the phosphosite in its 3D structure (if structure information/mapping is available).
- The color of every substrate has ben kept different in the Sunburst diagram for visual clarity.
- Clicking in the top right corner on the camara icon will download the image.
| Substrate | Organism | PSite | Sequence(+/-7) | Conservation | Disorder | Curator Assessment | Reliability | Evidence Class | Evidence Logic | PubMed | Phospho-ELM | PhosphoSite-Plus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SRF (P11831) | Homo sapiens | S85 | GSEGDSESGEEEELG | 0.877 | 0.7718 | - | - | - | 72 | 73 | 55 | 135 | | 10318869 | - | - |
| SRF (P11831) | Homo sapiens | S103 | RGLKRSLSEMEIGMV | 0.283 | 0.494 | TP | certain | experimental | 72 | 73 | 55 | 135 | | 10318869 | - | - |
| HNRNPA0 (Q13151) | Homo sapiens | S84 | VELKRAVSREDSARP | 0.157 | 0.5901 | - | - | - | 73 | 9 | 135 | 133 | 148 | 55 | 72 | | 12456657 | - | - |
| TH (P07101-2) | Homo sapiens | S67 | RFIGRRQSLIEDARK | 0.489 | 0.6183 | TP | certain | experimental | 73 | 107 | | 7901013 | - |
|
| EEF2K (O00418) | Homo sapiens | S377 | PPLLRPLSENSGDEN | 0.736 | 0.8565 | TP | certain | experimental | 135 | 133 | 73 | 55 | | 12171600 | Non high throughput (12171600) |
|
| TP53 (P04637) | Homo sapiens | S20 | PLSQETFSDLWKLLP | 0.567 | 0.6183 | TP | certain | experimental | 73 | 55 | | 11896587 | - |
|
| KRT18 (P05783) | Homo sapiens | S53 | ISVSRSTSFRGGMGS | 0.418 | 0.4476 | TP | certain | experimental | 73 | 55 | | 20724476 | - |
|
| TH (P07101) | Homo sapiens | S19 | KGFRRAVSELDAKQA | 0.455 | 0.5665 | TP | certain | experimental | 73 | 107 | | 7901013 | Non high throughput (7901013) |
|
| TH (P07101) | Homo sapiens | S71 | RFIGRRQSLIEDARK | 0.945 | 0.6183 | TP | certain | experimental | 73 | 107 | | 7901013 | - |
|
| TH (P07101-2) | Homo sapiens | S19 | KGFRRAVSELDAKQA | 0.455 | 0.5577 | TP | certain | experimental | 73 | 107 | | 7901013 | - |
|
| TH (P07101-3) | Homo sapiens | S19 | KGFRRAVSELDAKQA | 0.455 | 0.4801 | TP | certain | experimental | 73 | 107 | | 7901013 | - |
|
| TH (P07101-3) | Homo sapiens | S40 | RFIGRRQSLIEDARK | 0.090 | 0.5296 | TP | certain | experimental | 73 | 107 | | 7901013 | - |
|
| VIM (P08670) | Homo sapiens | S39 | TTSTRTYSLGSALRP | 0.934 | 0.4652 | TP | certain | experimental | 73 | 135 | 55 | 50 | | 12761892 | Non high throughput (12761892) |
|
| VIM (P08670) | Homo sapiens | S51 | LRPSTSRSLYASSPG | 0.118 | 0.4685 | TP | certain | experimental | 73 | 135 | 55 | 50 | | 12761892 | Non high throughput (12761892) |
|
| VIM (P08670) | Homo sapiens | S56 | SRSLYASSPGGVYAT | 0.931 | 0.5456 | TP | certain | experimental | 73 | 55 | 50 | | 12761892 | Non high throughput (12761892) |
|
| VIM (P08670) | Homo sapiens | S83 | GVRLLQDSVDFSLAD | 0.105 | 0.2849 | TP | certain | experimental | 73 | 55 | 50 | | 12761892 | Non high throughput (12761892) |
|
| Hspb1 (P14602) | Mus musculus | S15 | FSLLRSPSWEPFRDW | - | 0.1092 | TP | certain | experimental | 72 | 107 | | 1332886 | - |
|
| Hspb1 (P14602) | Mus musculus | S86 | RALNRQLSSGVSEIR | - | 0.3599 | TP | certain | experimental | 72 | 107 | | 1332886 | - |
|
| Lsp1 * (P19973 ) | Mus musculus | S195 | KLADRTESLNRSIKK | - | 0.7672 | U | likely | experimental | 73 | | 8995217 | - |
|
| Lsp1 * (P19973 ) | Mus musculus | S243 | PKLSRQPSIELPSMA | - | 0.6948 | U | likely | experimental | 73 | | 8995217 | - |
|
| Zfp36 (P22893) | Mus musculus | S316 | LPIFNRISVSE____ | - | 0.4766 | TP | certain | experimental | 73 | 214 | 72 | | 14688255 | Non high throughput (14688255) | - |
| Zfp36 * (P22893 ) | Mus musculus | S80 | PRPGPELSPSPTSPT | - | 0.8853 | U | likely | experimental | 72 | 214 | | 14688255 | Non high throughput (14688255) | - |
| Zfp36 * (P22893 ) | Mus musculus | S82 | PGPELSPSPTSPTAT | - | 0.9126 | U | likely | experimental | 72 | 214 | | 14688255 | Non high throughput (14688255) | - |
| Zfp36 * (P22893 ) | Mus musculus | S85 | ELSPSPTSPTATPTT | - | 0.9081 | U | likely | experimental | 72 | 214 | | 14688255 | Non high throughput (14688255) | - |
| Zfp36 * (P22893 ) | Mus musculus | T250 | PSCRRSTTPSTIWGP | - | 0.374 | U | likely | experimental | 73 | 214 | 72 | | 14688255 | Non high throughput (14688255) | - |
| LSP1 (P33241) | Homo sapiens | S204 | KLIDRTESLNRSIEK | 0.708 | 0.7289 | TP | likely | - | 14 | | 8995217 | Non high throughput (8995217) | - |
| LSP1 (P33241) | Homo sapiens | S252 | PKLARQASIELPSMA | 0.666 | 0.6136 | TP | likely | - | 14 | | 8995217 | Non high throughput (8995217) |
|
| Ddit3 (P35639) | Mus musculus | S78 | EVTRTSQSPRSPDSS | - | 0.9329 | TP | certain | - | 73 | 9 | 107 | | 8650547 | Non high throughput (8650547) | - |
| Ddit3 (P35639) | Mus musculus | S81 | RTSQSPRSPDSSQSS | - | 0.9013 | TP | certain | - | 73 | 9 | 107 | | 8650547 | Non high throughput (8650547) | - |
| VIM (P48616) | Bos taurus | S39 | TTSTRTYSLGSALRP | - | 0.4801 | TP | certain | experimental | 135 | 139 | 55 | 50 | 72 | 73 | | 12761892 | - |
|
| VIM (P48616) | Bos taurus | S56 | SRTLYTSSPGGVYAT | - | 0.6089 | TP | certain | experimental | 135 | 139 | 55 | 50 | 72 | 73 | | 12761892 | - |
|
| VIM (P48616) | Bos taurus | S83 | GVRLLQDSVDFSLAD | - | 0.2988 | TP | certain | experimental | 135 | 139 | 55 | 50 | 72 | 73 | | 12761892 | - |
|
| VIM (P48616) | Bos taurus | T51 | LRPTTSRTLYTSSPG | - | 0.5254 | TP | certain | experimental | 135 | 139 | 55 | 50 | 72 | 73 | | 12761892 | - |
|
| MAPKAPK2 * (P49137 ) | Homo sapiens | T338 | VPQTPLHTSRVLKED | 0.731 | 0.6427 | TP | likely | experimental | 73 | 148 | 28 | | 8846784 | Non high throughput (8846784) |
|
| MDM2 (Q00987) | Homo sapiens | S157 | SHLVSRPSTSSRRRA | 0.615 | 0.6375 | TP | certain | experimental | 72 | 73 | 55 | 9 | 273 | | 15688025 | - |
|
| MDM2 (Q00987) | Homo sapiens | S166 | SSRRRAISETEENSD | 0.815 | 0.7718 | TP | certain | experimental | 72 | 73 | 55 | 9 | 273 | | 15688025 | - |
|
| RCSD1 (Q6JBY9) | Homo sapiens | S179 | RRFRRSQSDCGELGD | 0.793 | 0.7547 | TP | certain | experimental | 214 | 135 | 55 | 73 | 148 | | 15850461 | - |
|
| RCSD1 (Q6JBY9) | Homo sapiens | S244 | PPLRRSPSRTEKQEE | 0.906 | 0.9308 | TP | likely | experimental | 14 | | 15850461 | - |
|
| Hnrnpa0 (Q9CX86) | Mus musculus | S84 | VELKRAVSREDSARP | - | 0.5901 | TP | certain | experimental | 148 | 55 | | 12456657 | Non high throughput (12456657) | - |
| Krt20 (Q9D312) | Mus musculus | S13 | QSFHRSLSSSSQGPA | - | 0.4582 | TP | - | - | - | - | - |
|
| HSPB1 (P04792) | Homo sapiens | S15 | FSLLRGPSWDPFRDW | 0.564 | 0.1323 | TP | certain | experimental | 18 | 72 | 62 | 50 | | 10383393 | - | - |
| HSPB1 (P04792) | Homo sapiens | S78 | PAYSRALSRQLSSGV | 0.601 | 0.3286 | TP | certain | experimental | 18 | 72 | 62 | 50 | | 10383393 | - | - |
| HSPB1 (P04792) | Homo sapiens | S82 | RALSRQLSSGVSEIR | 0.622 | 0.374 | TP | certain | experimental | 18 | 72 | 62 | 50 | | 10383393 | - | - |
| Zfp36 (P22893) | Mus musculus | S52 | RLTGRSTSLVEGRSC | - | 0.3392 | TP | certain | experimental | 72 | 73 | 45 | 9 | | 14688255 | - | - |
| Zfp36 (P22893) | Mus musculus | S178 | HVLRQSISFSGLPSG | - | 0.6375 | TP | certain | experimental | 72 | 73 | 45 | 9 | | 14688255 | - | - |
| TSC2 * (P49815 ) | Homo sapiens | S1254 | TALYKSLSVPAASTA | 0.263 | 0.5707 | TP | certain | experimental | 74 | 9 | 107 | 45 | | 12582162 | - | - |
| BAG2 (O95816) | Homo sapiens | S20 | GRFCRSSSMADRSSR | 0.053 | 0.4801 | TP | certain | experimental | 45 | 148 | 9 | 55 | 140 | | 15271996 | - | - |
| ALOX5 (P09917) | Homo sapiens | S272 | CSLERQLSLEQEVQQ | 0.144 | 0.1852 | TP | certain | experimental | 9 | 72 | 107 | | 11844797 | - | - |
| YWHAZ (P63104) | Homo sapiens | S58 | VVGARRSSWRVVSSI | 0.630 | 0.268 | TP | certain | experimental | 9 | 132 | 55 | | 12861023 | - | - |
| ARPC5 (O15511) | Homo sapiens | S77 | AVKDRAGSIVLKVLI | 0.096 | 0.1554 | TP | certain | experimental | 38 | 55 | 9 | | 12829704 | - | - |
| CDC25B (P30305) | Homo sapiens | S169 | VLRNITNSQAPDGRR | 0.502 | 0.7951 | TP | certain | experimental | 45 | 55 | 72 | | 16861915 | - | - |
| CDC25B (P30305) | Homo sapiens | S323 | QRLFRSPSMPCSVIR | 0.939 | 0.2817 | TP | certain | experimental | 55 | 72 | | 16861915 | - | - |
| CDC25B (P30305) | Homo sapiens | S353 | VQNKRRRSVTPPEEQ | 0.838 | 0.8713 | TP | certain | experimental | 45 | 55 | 72 | | 16861915 | - | - |
| CDC25B (P30305) | Homo sapiens | S375 | ARVLRSKSLCHDEIE | 0.895 | 0.3053 | TP | certain | experimental | 45 | 55 | 72 | | 16861915 | - | - |
| HSF1 (Q00613) | Homo sapiens | S121 | NIKRKVTSVSTLKSE | 0.090 | 0.2503 | TP | certain | experimental | 45 | 72 | 73 | 23 | 9 | 32 | | 16278218 | - | - |
| LIMK1 (P53667) | Homo sapiens | S323 | KDLGRSESLRVVCRP | 0.977 | 0.4087 | TP | certain | experimental | 9 | | 16456544 | - | - |
| AKT1 * (P31749 ) | Homo sapiens | S473 | RPHFPQFSYSASGTA | 0.952 | 0.5342 | TP | certain | experimental | 73 | 74 | 55 | 38 | 30 | | 11042204 | - | - |
| PLK1 * (P53350 ) | Homo sapiens | S326 | LTIPPRFSIAPSSLD | 0.181 | 0.2783 | TP | certain | experimental | 62 | 55 | 23 | | 18695677 | - | - |
| ZFP36L1 (Q07352) | Homo sapiens | S54 | GGFPRRHSVTLPSSK | 0.980 | 0.5854 | TP | likely | experimental | 72 | 9 | | 18326031 | - | - |
| ZFP36L1 (Q07352) | Homo sapiens | S92 | RFRDRSFSEGGERLL | 0.965 | 0.5533 | TP | certain | experimental | 72 | 9 | 45 | | 18326031 | - | - |
| ZFP36L1 (Q07352) | Homo sapiens | S203 | PRLQHSFSFAGFPSA | 0.946 | 0.3983 | TP | certain | experimental | 72 | 9 | 45 | | 18326031 | - | - |
| Etv1 (P41164) | Mus musculus | S216 | PMYQRQMSEPNIPFP | - | 0.8313 | TP | certain | experimental | 72 | 73 | 9 | | 11551945 | - | - |
| Etv1 (P41164) | Mus musculus | S191 | HRFRRQLSEPCNSFP | - | 0.6089 | TP | certain | experimental | 72 | 73 | 9 | | 11551945 | - | - |
| RTN4 (Q9NQC3-2) | Homo sapiens | S107 | VAPERQPSWDPSPVS | 0.000 | 0.8655 | TP | certain | experimental | 72 | 9 | 135 | 140 | | 16095439 | - | - |
| TRIM29 (Q14134) | Homo sapiens | S550 | GYPSLMRSQSPKAQP | 0.502 | 0.5456 | TP | certain | experimental | 72 | 9 | 148 | | 24469230 | - | - |
| TRIM29 * (Q14134 ) | Homo sapiens | S552 | PSLMRSQSPKAQPQT | 0.372 | 0.6136 | TP | certain | experimental | 72 | 9 | 148 | | 24469230 | - | - |
| PIAS1 * (O75925 ) | Homo sapiens | S522 | DTSYINTSLIQDYRH | 0.144 | 0.391 | TP | certain | experimental | 72 | 8 | 140 | | 23202365 | - | - |
| AATF (Q9NY61) | Homo sapiens | T366 | FTVYRNRTLQKWHDK | 0.687 | 0.2258 | TP | certain | experimental | 45 | 72 | 9 | | 22909821 | - | - |
| TH (P07101) | Homo sapiens | S40 | GQGAPGPSLTGSPWP | 0.090 | 0.7843 | TP | certain | experimental | 73 | 107 | | 7901013 | Non high throughput (7901013) | - |
| CREB1 (P16220) | Homo sapiens | S133 | EILSRRPSYRKILND | - | 0.4979 | - | - | - | - | 8887554 | Non high throughput (8887554) |
|
| AGO2 (Q9UKV8) | Homo sapiens | S387 | SKLMRSASFNTDPYV | 0.070 | 0.3053 | - | - | - | - | - | - |
|
| PARN (O95453) | Homo sapiens | S557 | NHYYRNNSFTAPSTV | 0.510 | 0.4541 | - | - | - | - | - | - |
|
| CRYAB * (P02511 ) | Homo sapiens | S59 | PSFLRAPSWFDTGLS | 0.186 | 0.2258 | TP | certain | experimental | 72 | 55 | | 9774459 | - | - |
| TH (P07101-3) | Homo sapiens | S404 | GELLHCLSEEPEIRA | 0.770 | 0.2292 | - | - | - | - | - | - |
|
| Creb1 (P15337) | Rattus norvegicus | S133 | EILSRRPSYRKILND | - | 0.4979 | TP | certain | experimental | 73 | 55 | 148 | | 8887554 | - |
|
| Rps6ka1 (P18653) | Mus musculus | S369 | HQLFRGFSFVATGLM | - | 0.2002 | - | - | - | - | - | - |
|
| ATF1 (P18846) | Homo sapiens | S63 | GILARRPSYRKILKD | 0.920 | 0.4864 | - | - | - | - | - | - | - |
| SHC1 (P29353) | Homo sapiens | S17 | YNPLRNESLSSLEEG | 0.950 | 0.7843 | - | - | - | - | - | - | - |
| CDC25B (P30305-2) | Homo sapiens | S309 | QRLFRSPSMPCSVIR | 0.271 | 0.2817 | - | - | - | - | - | - | - |
| CDC25C (P30307) | Homo sapiens | S216 | SGLYRSPSMPENLNR | 0.868 | 0.6136 | TP | certain | predicted | 14 | | 15629715 | - |
|
| KRT20 (P35900) | Homo sapiens | S13 | RSFHRSLSSSLQAPV | 0.855 | 0.3807 | - | - | - | - | - | - |
|
| PLK1 (P53350) | Homo sapiens | S383 | DMLQQLHSVNASKPS | 0.446 | 0.5055 | U | likely | predicted | 14 | | 18695677 | - |
|
| Pde4a (P54748) | Rattus norvegicus | S147 | SFLYRSDSDYDMSPK | - | 0.5055 | - | - | - | - | - | - |
|
| Creb1 * (Q01147 ) | Mus musculus | S133 | EILSRRPSYRKILND | - | 0.4979 | TP | likely | experimental | 73 | | 10679065 | - |
|
| BCL2L1 * (Q07817 ) | Homo sapiens | S62 | PSWHLADSPAVNGAT | 0.275 | 0.6089 | U | likely | experimental | 73 | | 24621501 | - |
|
| Lpcat2 (Q8BYI6) | Mus musculus | S34 | PMVPRQASFFPPPVP | - | 0.2783 | - | - | - | - | - | - |
|
| Gys1 (Q9Z1E4) | Mus musculus | S8 | MPLSRSLSVSSLPGL | - | 0.2609 | - | - | - | - | - | - | - |
| HSF1 (Q00613) | Homo sapiens | T120 | ENIKRKVTSVSTLKS | 0.684 | 0.2541 | - | - | - | - | - | - | - |
| Rps6ka3 (P18654) | Mus musculus | S227 | DHEKKAYSFCGTVEY | - | 0.1852 | - | - | - | 72 | 55 | | - | - |
|
| WNK4 (Q96J92) | Homo sapiens | S575 | PFLFRHASYSSTTSD | 0.789 | 0.5342 | TP | certain | experimental | 73 | 74 | 8 | 55 | | 26732173 | - |
|
| Rbm7 (Q9CQT2) | Mus musculus | S136 | QMVQRSFSSPEDYQR | - | 0.6136 | TP | certain | - | 135 | 73 | 72 | 214 | 8 | 148 | 55 | | 25525152 | - |
|
| MRTFA (Q969V6) | Homo sapiens | S333 | CGLARQNSTSLTGKP | 0.655 | 0.662 | TP | certain | - | 73 | 9 | 72 | 55 | 148 | | 27492266 | - |
|
| Ripk1 (Q60855) | Mus musculus | S336 | LPPSRSNSEQPGSLH | - | 0.7036 | TP | certain | - | 72 | 3 | 214 | 148 | 8 | 73 | 23 | | 28920954 | - |
|
| TH (P07101-4) | Homo sapiens | S44 | RFIGRRQSLIEDARK | 0.066 | 0.5758 | - | - | - | - | - | - |
|
| Mrtfa (Q8K4J6) | Mus musculus | S371 | SGLARQSSTALAAKP | - | 0.662 | TP | certain | experimental | 72 | 9 | | 27492266 | - |
|
| CEP131 (Q9UPN4) | Homo sapiens | S47 | KPIVRSVSVVTGSEQ | 0.844 | 0.7036 | TP | certain | experimental | 214 | 55 | 73 | 9 | | 26616734 | - |
|
| Mrtfa (Q8K4J6) | Mus musculus | S351 | PTPSRSLSTSSSPSS | - | 0.919 | TP | certain | experimental | 72 | 9 | | 27492266 | - |
|
| DDX5 (P17844) | Homo sapiens | S198 | CRACRLKSTCIYGGA | 0.065 | 0.1028 | TP | certain | experimental | 72 | 9 | 214 | 73 | 55 | 28 | 23 | 143 | | 23482664 | - |
|
| CEP131 (Q9UPN4) | Homo sapiens | S78 | NNLRRSNSTTQVSQP | 0.950 | 0.8462 | TP | certain | experimental | 214 | 55 | 73 | 9 | | 26616734 | - |
|
| DAZL (Q92904) | Homo sapiens | S65 | SFFARYGSVKEVKII | 0.026 | 0.2193 | TP | certain | experimental | 73 | 9 | 55 | 148 | | 27280388 | - |
|
| RIPK1 (Q13546) | Homo sapiens | S320 | AVVKRMQSLQLDCVA | 0.919 | 0.384 | TP | certain | experimental | 73 | 55 | 148 | | 28506461 | - |
|
| Hif3a (Q0VBL6-2) | Mus musculus | S184 | HFSLRMKSTLTSRGR | - | 0.5139 | TP | certain | experimental | 214 | 72 | 8 | | 29054108 | - |
|
| TH (P07101-4) | Homo sapiens | S19 | KGFRRAVSELDAKQA | 0.455 | 0.4766 | - | - | - | - | - | - |
|
| TRIM28 (Q13263) | Homo sapiens | S473 | SGVKRSRSGEGEVSG | 0.459 | 0.7163 | TP | certain | experimental | 73 | 9 | 148 | 72 | | 23740979 | - |
|
| MRTFA (Q969V6) | Homo sapiens | S312 | TPPVRSLSTTNSSSS | 0.331 | 0.8343 | TP | certain | experimental | 73 | 9 | 72 | 55 | 148 | | 27492266 | - |
|
| PFKFB3 (Q16875) | Homo sapiens | S461 | NPLMRRNSVTPLASP | 0.121 | 0.7595 | TP | certain | experimental | 148 | 73 | 214 | 72 | 8 | 55 | | 23548149 | - |
|
| Rps6ka3 (P18654) | Mus musculus | S386 | HQLFRGFSFVAITSD | - | 0.247 | TP | certain | experimental | 73 | 55 | 28 | 148 | | 17906627 | - |
|
| BECN1 (Q14457) | Homo sapiens | S90 | IPPARMMSTESANSF | 0.467 | 0.4582 | TP | certain | experimental | 72 | 73 | 74 | 8 | 55 | | 25693418 | - |
|
| Ripk1 (Q60855) | Mus musculus | S321 | PVLQRMFSLQHDCVP | - | 0.3983 | TP | certain | experimental | 72 | 3 | 214 | 148 | 8 | 73 | 23 | | 28920954 | - |
|
| ZFP36 (P26651) | Homo sapiens | S113 | TELCRTFSESGRCRY | 0.130 | 0.3117 | - | - | - | - | - | - | - |
| ZFP36 (P26651) | Homo sapiens | S184 | HPPVLRQSISFSGLP | 0.952 | 0.6136 | - | - | - | - | - | - | - |
| ZFP36 (P26651) | Homo sapiens | S186 | PVLRQSISFSGLPSG | 0.961 | 0.6322 | TP | certain | - | 72 | 8 | 143 | 55 | | 15944294 | - | - |
| ZFP36 (P26651) | Homo sapiens | S273 | GGLVRTPSVQSLGSD | 0.963 | 0.8013 | - | - | - | - | - | - | - |
| ZFP36 (P26651) | Homo sapiens | S323 | LPIFNRISVSE____ | 0.931 | 0.3774 | - | - | - | - | - | - | - |
| ZFP36 (P26651) | Homo sapiens | S60 | RLPGRSTSLVEGRSC | 0.751 | 0.3807 | TP | certain | experimental | 72 | 8 | 143 | 55 | | 15944294 | - | - |
| ZFP36 (P26651) | Homo sapiens | S66 | TSLVEGRSCGWVPPP | 0.285 | 0.4582 | U | likely | predicted | 14 | | 20221403 | - | - |
| PDE4A (P27815) | Homo sapiens | S152 | SFLYRSDSDYDMSPK | 0.584 | 0.562 | TP | certain | experimental | 72 | 73 | 8 | 55 | 148 | | 21323643 | - | - |
| LSP1 (P33241-2) | Homo sapiens | S252 | STIKSTPSGKRYKFV | 0.666 | 0.4979 | - | - | - | - | - | - | - |
| ETV1 (P50549) | Homo sapiens | S191 | HRFRRQLSEPCNSFP | 0.258 | 0.6043 | TP | certain | experimental | 72 | 73 | 55 | 8 | | 11551945 | - | - |
| RPS6KA3 (P51812) | Homo sapiens | S386 | HQLFRGFSFVAITSD | 0.951 | 0.247 | - | - | - | - | - | - | - |
| E2F1 (Q01094) | Homo sapiens | S364 | PLLSRMGSLRAPVDE | 0.364 | 0.6322 | TP | certain | experimental | 72 | 73 | 8 | 55 | | 22802261 | - | - |
| RPS6KA1 (Q15418) | Homo sapiens | S380 | HQLFRGFSFVATGLM | 0.945 | 0.2094 | - | - | - | - | - | - | - |
| LPCAT2 (Q7L5N7) | Homo sapiens | S34 | PMVPRQASFFPPPVP | 0.920 | 0.2541 | TP | certain | predicted | 14 | | 20663880 | - | - |
| TAB3 * (Q8N5C8 ) | Homo sapiens | S506 | HKYQRSSSSGSDDYA | 0.777 | 0.6661 | U | likely | experimental | 72 | 28 | | 18021073 | - | - |
| MYLK2 (Q9H1R3) | Homo sapiens | S151 | PAFLHSPSCPAIISS | 0.943 | 0.7163 | - | - | - | - | - | - | - |
| MYLK2 * (P07313 ) | Oryctolagus cuniculus | S161 | PAFLHSPSCPAIIAS | - | 0.6991 | U | likely | experimental | 73 | 135 | 133 | | 11795875 | - | - |
| RTN4 (Q9NQC3-2) | Homo sapiens | S7 | _MEDLDQSPLVSSSD | 0.000 | 0.9621 | - | - | - | - | - | - | - |
| RPS6KA1 (E9PGT3) | Homo sapiens | S369 | HQLFRGFSFVATGLM | - | 0.2094 | - | - | - | - | - | - | - |
| ZFP36 (M0R0H3) | Homo sapiens | S203 | PVLRQSISFSGLPSG | - | 0.6322 | - | - | - | - | - | - | - |
| ZFP36 (M0R0H3) | Homo sapiens | S340 | LPIFNRISVSE____ | - | 0.3774 | - | - | - | - | - | - | - |
| ZFP36 (M0R0H3) | Homo sapiens | S77 | RLPGRSTSLVEGRSC | - | 0.4017 | - | - | - | - | - | - | - |
| ZFP36 (M0R0H3) | Homo sapiens | S105 | PRLGPELSPSPTSPT | - | 0.8162 | - | - | - | - | - | - | - |
| ZFP36 (M0R0H3) | Homo sapiens | S107 | LGPELSPSPTSPTAT | - | 0.8565 | - | - | - | - | - | - | - |
| ZFP36 (M0R0H3) | Homo sapiens | S110 | ELSPSPTSPTATSTT | - | 0.8655 | - | - | - | - | - | - | - |
| DDIT3 (Q53YD1) | Homo sapiens | S79 | EVTSTSQSPHSPDSS | - | 0.9519 | - | - | - | - | - | - | - |
| DDIT3 (Q53YD1) | Homo sapiens | S82 | STSQSPHSPDSSQSS | - | 0.9039 | - | - | - | - | - | - | - |
| (A8K6K7) | Homo sapiens | S8 | MPLNRTLSMSSLPGL | - | 0.2817 | - | - | - | - | - | - | - |
| ETV1 (P50549) | Homo sapiens | S216 | PMYQRQMSEPNIPFP | 0.364 | 0.8313 | TP | certain | - | 72 | 73 | 55 | 8 | | 11551945 | - | - |
| (A8K2L4) | Homo sapiens | S204 | KLIDRTESLNRSIEK | - | 0.7289 | - | - | - | - | - | - | - |
| RPS6KA3 (P51812) | Homo sapiens | S227 | DHEKKAYSFCGTVEY | 0.758 | 0.1852 | - | - | - | - | - | - | - |
| RBM7 (Q6IRX3) | Homo sapiens | S136 | QIIQRSFSSPENFQR | - | 0.5211 | TP | certain | - | 72 | 73 | 8 | 55 | | 25525152 | - | - |
| RIPK1 (A0A024QZU0) | Homo sapiens | S320 | AVVKRMQSLQLDCVA | - | 0.384 | - | - | - | - | - | - | - |
| PDE4A (P27815-7) | Homo sapiens | S130 | SFLYRSDSDYDMSPK | 0.792 | 0.5577 | - | - | - | - | - | - | - |
| LSP1 (P33241-2) | Homo sapiens | S142 | KLIDRTESLNRSIEK | 0.072 | 0.7289 | - | - | - | - | - | - | - |
| ZFP36 (P26651) | Homo sapiens | T257 | CPSCRRATPISVWGP | 0.223 | 0.2951 | - | - | - | - | - | - | - |
| ZFP36 (P26651) | Homo sapiens | T271 | PLGGLVRTPSVQSLG | 0.060 | 0.562 | - | - | - | - | - | - | - |
| ZFP36 (M0R0H3) | Homo sapiens | T274 | CPSCRRATPISVWGP | - | 0.2951 | - | - | - | - | - | - | - |
Help Instructions
- Hovering on the "*" in "Substrate" column will display the text if there is a note by the annotator for the given site.
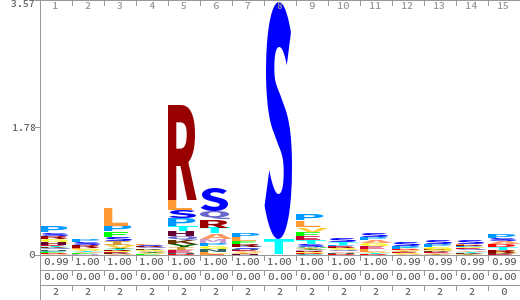
- The column having "Sequence" motif represents phosphosite and its neighbouring 7 residues (upstream and downstream).
- The score in "Conservation" column varies from 0 to 1; Higher the score more conserved is the site.
- The score in "Disorder" column varies from 0 to 1; Score higher than 0.4 suggests disorder.
- In the "Curator Assessment" column, TP refers to True Positive; FP refers to False Positive; and U refers to unknown if the annoatator cannot decide the status based on available information.
- Hovering on the number in the "Evidence Logic" column will show the name of method used to characterize/infer the phosphosite (for more details plase follow the link)