Help Instruction
- Double clicking on the substrate will redirect to the uniprot entry.
- Clicking in the block having the substrate will zoom into its phophosites
- Clicking the center of the circle will lead to zooming out.
- Clicking on the phosphosite will render the phosphosite in its 3D structure (if structure information/mapping is available).
- The color of every substrate has ben kept different in the Sunburst diagram for visual clarity.
- Clicking in the top right corner on the camara icon will download the image.
| Substrate | Organism | PSite | Sequence(+/-7) | Conservation | Disorder | Curator Assessment | Reliability | Evidence Class | Evidence Logic | PubMed | Phospho-ELM | PhosphoSite-Plus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PRKAB1 (Q9Y478) | Homo sapiens | S108 | SKLPLTRSHNNFVAI | 0.861 | 0.2752 | TP | certain | experimental | 214 | 73 | | 19376078 | - |
|
| PRKAB1 (Q9Y478) | Homo sapiens | S174 | MVDSQKCSDVSELSS | 0.220 | 0.433 | TP | certain | experimental | 214 | 73 | | 19376078 | - |
|
| PRKAB1 (Q9Y478) | Homo sapiens | S177 | SQKCSDVSELSSSPP | 0.214 | 0.4582 | TP | certain | experimental | 214 | 73 | | 19376078 | - |
|
| PRKAB1 (Q9Y478) | Homo sapiens | S24 | HKTPRRDSSGGTKDG | 0.763 | 0.9308 | TP | certain | experimental | 214 | 73 | | 19376078 | - |
|
| PRKAB1 (Q9Y478) | Homo sapiens | T158 | NIIQVKKTDFEVFDA | 0.163 | 0.1184 | TP | certain | experimental | 214 | 73 | | 19376078 | - |
|
| PRKAB1 (Q9Y478) | Homo sapiens | T80 | APAQARPTVFRWTGG | 0.836 | 0.6531 | TP | certain | experimental | 214 | 73 | | 19376078 | - |
|
| PPP1R3C (Q9UQK1) | Homo sapiens | S293 | LESTIFGSPRLASGL | 0.839 | 0.3087 | TP | certain | experimental | 23 | 72 | 214 | 9 | | 19171932 | - |
|
| PPP1R3C (Q9UQK1) | Homo sapiens | S33 | MRLCLAHSPPVKSFL | 0.979 | 0.1251 | TP | certain | experimental | 23 | 72 | 214 | 9 | | 19171932 | - |
|
| Slc12a1 (P55014) | Mus musculus | S126 | GPKVNRPSLLEIHEQ | - | 0.4051 | TP | certain | experimental | 23 | 73 | 107 | 9 | 55 | 72 | | 17341212 | - |
|
| RRN3 (Q9NYV6) | Homo sapiens | S635 | DTHFRSPSSSVGSPP | 0.237 | 0.4017 | TP | certain | experimental | 23 | 73 | 9 | 72 | 55 | | 19815529 | - |
|
| ACACA (Q13085) | Homo sapiens | S80 | LHIRSSMSGLHLVKQ | 0.956 | 0.2849 | TP | certain | not annotated | 72 | 214 | 55 | 74 | | 26456332 | - |
|
| KLC2 (Q9H0B6) | Homo sapiens | S545 | GSLRRSGSFGKLRDA | 0.830 | 0.6136 | TP | certain | experimental | 73 | 214 | 135 | 55 | 9 | | 21725060 | - |
|
| KLC2 (Q9H0B6) | Homo sapiens | S582 | PRMKRASSLNFLNKS | 0.773 | 0.8013 | TP | certain | experimental | 73 | 214 | 55 | 135 | 9 | | 21725060 | - |
|
| Gabbr1 (Q9Z0U4) | Rattus norvegicus | S948 | ELRHQLQSRQQLRSR | - | 0.6322 | TP | certain | experimental | 73 | 23 | 135 | 9 | | 17224405 | - |
|
| Mlxipl (Q8VIP2) | Rattus norvegicus | S568 | TLLRPPESPDAVPEI | - | 0.7799 | TP | certain | experimental | 73 | 62 | | 11724780 | - |
|
| Mtor * (Q9JLN9 ) | Mus musculus | T2446 | NKRSRTRTDSYSAGQ | - | 0.662 | TP | certain | experimental | 73 | 8 | | 14970221 | - |
|
| ZNF692 (Q9BU19) | Homo sapiens | S470 | VAAHRSKSHPALLLA | 0.945 | 0.5854 | TP | certain | experimental | 73 | 8 | | 17097062 | - |
|
| EEF2K (O00418) | Homo sapiens | S398 | DSLPSSPSSATPHSQ | 0.546 | 0.7163 | TP | certain | experimental | 72 | 73 | 55 | 214 | 8 | 107 | | 14709557 | - |
|
| ACACB * (O00763 ) | Homo sapiens | S222 | PTMRPSMSGLHLVKR | 0.966 | 0.6043 | TP | likely | experimental | 55 | | 12488245 | - |
|
| ULK1 (O75385) | Homo sapiens | S638 | FDFPKTPSSQNLLAL | 0.789 | 0.4864 | - | - | experimental | 72 | 214 | | 21205641 | - |
|
| Gabbr2 (O88871) | Rattus norvegicus | S783 | VTSVNQASTSRLEGL | - | 0.5017 | TP | certain | experimental | 66 | 73 | 31 | 8 | 72 | 55 | | 17224405 | - |
|
| Mylk (P11799) | Gallus gallus | S1749 | RAIGRLSSMAMISGM | - | 0.3704 | TP | certain | experimental | 73 | 31 | | 18426792 | - |
|
| Rb1 (P13405) | Mus musculus | S804 | IYISPLKSPYKISEG | - | 0.3182 | TP | certain | experimental | 73 | 73 | 148 | 8 | 55 | | 19217427 | - |
|
| TNNI3 (P19429) | Homo sapiens | S150 | TLRRVRISADAMMQA | 0.393 | 0.384 | TP | certain | experimental | 72 | 73 | 55 | | 22493448 | - |
|
| NOS3 * (P29473 ) | Bos taurus | S1179 | TSRIRTQSFSLQERH | - | 0.4409 | TP | likely | experimental | 72 | 55 | 36 | | 12107173 | - |
|
| NOS3 (P29474) | Homo sapiens | S1177 | TSRIRTQSFSLQERQ | 0.140 | 0.433 | TP | certain | experimental | 72 | 55 | | 16126727 | - |
|
| CSNK1E (P49674) | Homo sapiens | S389 | RGAPANVSSSDLTGR | 0.200 | 0.7843 | TP | likely | experimental | 72 | 73 | 8 | | 17525164 | - |
|
| Tsc2 (P49816-2) | Rattus norvegicus | S1346 | QPLSKSSSSPELQTL | - | 0.8343 | TP | certain | experimental | 23 | 148 | 107 | 8 | 73 | 72 | | 14651849 | - |
|
| Kcnj11 (P70673) | Rattus norvegicus | S385 | AKPKFSISPDSLS__ | - | 0.3053 | TP | certain | experimental | 55 | 148 | 140 | 72 | | 19357830 | - |
|
| Cry1 (P97784) | Mus musculus | S71 | ANLRKLNSRLFVIRG | - | 0.1251 | TP | certain | experimental | 55 | 72 | 73 | 8 | | 19833968 | - |
|
| GFPT1 (Q06210-2) | Homo sapiens | S243 | CNLSRVDSTTCLFPV | 0.024 | 0.3087 | TP | certain | experimental | 72 | 73 | 55 | 9 | 214 | 107 | | 28008135 | - |
|
| Kcnma1 (Q08460-4) | Mus musculus | S722 | GRSERDCSCMSGRVR | - | 0.1791 | TP | likely | experimental | 73 | 9 | | 21209098 | - |
|
| PPP2R5C (Q13362) | Homo sapiens | S298 | KYWPKTHSPKEVMFL | 0.763 | 0.0349 | TP | certain | experimental | 73 | 9 | | 19366811 | - |
|
| PPP2R5C (Q13362) | Homo sapiens | S336 | RQLAKCVSSPHFQVA | 0.594 | 0.0888 | TP | certain | experimental | 73 | 9 | | 19366811 | - |
|
| PFKFB3 (Q16875) | Homo sapiens | S461 | NPLMRRNSVTPLASP | 0.121 | 0.7595 | TP | certain | experimental | 73 | 55 | | 12065600 | - |
|
| TBC1D1 (Q86TI0) | Homo sapiens | S237 | RPMRKSFSQPGLRSL | 0.733 | 0.422 | TP | certain | experimental | 55 | 8 | 73 | 214 | | 17995453 | - |
|
| Cdkn1b (P46414) | Mus musculus | T197 | KPGLRRQT_______ | - | 0.9473 | TP | certain | experimental | 73 | 8 | | 18701472 | - |
|
| CDKN1B (P46527) | Homo sapiens | T198 | PGLRRRQT_______ | 0.507 | 0.9488 | TP | certain | experimental | 72 | 73 | 8 | 55 | 273 | 107 | | 17237771 | - |
|
| Tsc2 (P49816-2) | Rattus norvegicus | T1228 | PTLPRSNTVASFSSL | - | 0.5176 | TP | certain | experimental | 23 | 148 | 107 | 8 | 72 | 73 | | 14651849 | - |
|
| VASP (P50552) | Homo sapiens | T278 | LARRRKATQVGEKTP | 0.261 | 0.8828 | TP | certain | experimental | 72 | 55 | 148 | 73 | 8 | | 17082196 | - |
|
| TBC1D1 (Q86TI0) | Homo sapiens | T596 | AFRRRANTLSHFPIE | 0.867 | 0.5419 | TP | certain | experimental | 55 | 73 | 214 | | 17995453 | - |
|
| Med1 (Q925J9-4) | Mus musculus | S656 | SPLERQNSSSGSPRM | - | 0.7951 | TP | certain | experimental | 72 | 73 | 9 | 31 | 23 | 55 | | 23943624 | - | - |
| FOXO3 * (O43524 ) | Homo sapiens | S399 | DNITLPPSQPSPTGG | 0.301 | 0.7672 | U | likely | experimental | 72 | 9 | | 17711846 | - | - |
| FOXO3 * (O43524 ) | Homo sapiens | S413 | GLMQRSSSFPYTTKG | 0.437 | 0.6227 | TP | certain | experimental | 72 | 9 | 31 | 55 | 73 | | 17711846 | - | - |
| FOXO3 * (O43524 ) | Homo sapiens | S555 | RALSNSVSNMGLSES | 0.204 | 0.5419 | U | likely | experimental | 72 | 9 | 31 | | 17711846 | - | - |
| FOXO3 * (O43524 ) | Homo sapiens | S588 | QTLSDSLSGSSLYST | 0.279 | 0.494 | U | likely | experimental | 72 | 9 | 55 | 73 | | 17711846 | - | - |
| FOXO3 * (O43524 ) | Homo sapiens | S626 | SLECDMESIIRSELM | 0.672 | 0.3529 | U | likely | experimental | 72 | 9 | 31 | 73 | | 17711846 | - | - |
| EP300 (Q09472) | Homo sapiens | S89 | SELLRSGSSPNLNMG | 0.391 | 0.6851 | TP | likely | experimental | 72 | 9 | 73 | | 11518699 | - | - |
| HDAC5 (Q9UQL6) | Homo sapiens | S259 | FPLRKTASEPNLKVR | 0.835 | 0.6712 | TP | certain | experimental | 9 | 23 | 3 | 72 | 55 | | 18184930 | - | - |
| HDAC5 (Q9UQL6) | Homo sapiens | S498 | RPLSRTQSSPLPQSP | 0.940 | 0.9126 | TP | certain | experimental | 9 | 23 | 3 | 72 | 55 | | 18184930 | - | - |
| Kcnb1 (P15387) | Rattus norvegicus | S444 | ERAKRNGSIVSMNMK | - | 0.3631 | TP | certain | experimental | 31 | 55 | 72 | 9 | 147 | 74 | | 22006306 | - | - |
| Kcnb1 (P15387) | Rattus norvegicus | S541 | SKMAKTQSQPILNTK | - | 0.6227 | TP | certain | experimental | 31 | 55 | 72 | 9 | 147 | 74 | | 22006306 | - | - |
| CFTR (P13569) | Homo sapiens | S737 | EPLERRLSLVPDSEQ | 0.978 | 0.6991 | TP | certain | experimental | 72 | 9 | 107 | | 19095655 | - | - |
| CFTR (P13569) | Homo sapiens | S768 | LQARRRQSVLNLMTH | 0.833 | 0.4369 | TP | certain | experimental | 72 | 9 | 107 | | 19095655 | - | - |
| CTBP1 (Q13363) | Homo sapiens | S158 | REGTRVQSVEQIREV | 0.085 | 0.3321 | TP | certain | experimental | 23 | 72 | 30 | 55 | | 23291169 | - | - |
| KPNA2 (P52292) | Homo sapiens | S105 | QAARKLLSREKQPPI | 0.931 | 0.5456 | TP | certain | experimental | 72 | 73 | 20 | 9 | | 15342649 | - | - |
| FOXO3 (O43524) | Homo sapiens | T179 | SSPDKRLTLSQIYEW | 0.645 | 0.3807 | TP | likely | experimental | 72 | 9 | | 17711846 | - | - |
| GBF1 (Q92538) | Homo sapiens | T1337 | GKIHRSATDADVVNS | 0.284 | 0.4507 | TP | certain | experimental | 73 | 55 | 9 | 20 | 72 | | 18063581 | - | - |
| PFKFB2 (O60825) | Homo sapiens | S466 | PVRMRRNSFTPLSSS | 0.089 | 0.5807 | - | - | - | - | - | - |
|
| Ulk1 (O70405) | Mus musculus | S467 | SAIRRSGSTTPLGFG | - | 0.7415 | - | - | - | - | - | - |
|
| Ulk1 (O70405) | Mus musculus | S555 | GLGCRLHSAPNLSDF | - | 0.5176 | - | - | - | - | - | - |
|
| ULK1 (O75385) | Homo sapiens | S638 | FDFPKTPSSQNLLAL | 0.789 | 0.4864 | - | - | - | - | - | - |
|
| TP53 (P04637) | Homo sapiens | S20 | PLSQETFSDLWKLLP | 0.567 | 0.6183 | - | - | - | - | - | - |
|
| Lipe (P15304) | Rattus norvegicus | S865 | ESMRRSVSEAALAQP | - | 0.5807 | - | - | - | - | - | - |
|
| NOS3 (P29474) | Homo sapiens | S633 | WRRKRKESSNTDSAG | 0.163 | 0.4725 | - | - | - | - | - | - |
|
| HNF4A (P41235) | Homo sapiens | S313 | GKIKRLRSQVQVSLE | 0.060 | 0.2364 | - | - | - | - | - | - |
|
| Tsc2 (P49816) | Rattus norvegicus | S1389 | QPLSKSSSSPELQTL | - | 0.8343 | - | - | - | - | - | - |
|
| LIPE (Q05469) | Homo sapiens | S855 | EPMRRSVSEAALAQP | 0.756 | 0.7415 | - | - | - | - | - | - |
|
| GFPT1 (Q06210) | Homo sapiens | S261 | CNLSRVDSTTCLFPV | 0.557 | 0.3149 | - | - | - | - | - | - |
|
| KLC1 (Q07866) | Homo sapiens | S521 | ENMEKRRSRESLNVD | 0.982 | 0.6806 | - | - | - | - | - | - |
|
| PRKAA1 (Q13131) | Homo sapiens | S360 | LATSPPDSFLDDHHL | 0.163 | 0.3494 | - | - | - | - | - | - |
|
| PRKAA1 (Q13131) | Homo sapiens | S486 | DEITEAKSGTATPQR | 0.159 | 0.6906 | - | - | - | - | - | - |
|
| PRKAA1 (Q13131) | Homo sapiens | S494 | GTATPQRSGSVSNYR | 0.112 | 0.5758 | - | - | - | - | - | - |
|
| PRKAA1 (Q13131) | Homo sapiens | S496 | ATPQRSGSVSNYRSC | 0.230 | 0.6043 | - | - | - | - | - | - |
|
| SLC12A1 (Q13621) | Homo sapiens | S130 | GPKVNRPSLLEIHEQ | 0.603 | 0.3807 | - | - | - | - | - | - |
|
| RPTOR (Q8N122) | Homo sapiens | S792 | DKMRRASSYSSLNSL | 0.494 | 0.3631 | - | - | - | - | - | - |
|
| TP53 (P04637) | Homo sapiens | T18 | EPPLSQETFSDLWKL | 0.691 | 0.6183 | - | - | - | - | - | - |
|
| PRKAA1 (Q13131) | Homo sapiens | T183 | SDGEFLRTSCGSPNY | 0.698 | 0.2884 | - | - | - | - | - | - |
|
| PRKAA1 (Q13131) | Homo sapiens | T388 | ETPRARHTLDELNPQ | 0.221 | 0.5577 | - | - | - | - | - | - |
|
| ATG9A (Q7Z3C6) | Homo sapiens | S761 | QSIPRSASYPCAAPR | 0.948 | 0.7799 | - | - | - | - | - | - |
|
| Nfe2l2 (Q60795) | Mus musculus | S550 | HLLKRRLSTLYLEVF | - | 0.1251 | - | - | - | - | - | - |
|
| Pde4b (Q8VBU5) | Mus musculus | S304 | KKLMHSSSLNNTSIS | - | 0.3529 | - | - | - | - | - | - |
|
| Foxo3 (Q9WVH4) | Mus musculus | S412 | GLMQRGSSFPYTAKS | - | 0.6227 | - | - | - | - | - | - |
|
| NET1 (Q7Z628) | Homo sapiens | S100 | RPLARVTSLANLISP | 0.976 | 0.4541 | - | - | - | - | - | - |
|
| FH (P07954) | Homo sapiens | S75 | YGAQTVRSTMNFKIG | 0.781 | 0.2575 | - | - | - | - | - | - |
|
| Tnni3 (P48787) | Mus musculus | S151 | TLRRVRISADAMMQA | - | 0.3948 | - | - | - | - | - | - |
|
| SNX17 (Q15036) | Homo sapiens | S437 | VKLSSKLSAVSLRGI | 0.508 | 0.3566 | - | - | - | - | - | - |
|
| GAPDH (P04406) | Homo sapiens | S122 | GAKRVIISAPSADAP | 0.602 | 0.1942 | - | - | - | - | - | - |
|
| JAK1 (P23458) | Homo sapiens | S518 | GSDRSFPSLGDLMSH | 0.880 | 0.3529 | - | - | - | - | - | - |
|
| YAP1 (P46937) | Homo sapiens | S61 | IVHVRGDSETDLEAL | 0.807 | 0.7916 | - | - | - | - | - | - |
|
| EPM2A (O95278) | Homo sapiens | S25 | PELLVVGSRPELGRW | 0.944 | 0.4256 | - | - | - | - | - | - |
|
| Sqstm1 (Q64337) | Mus musculus | S351 | SSKEVDPSTGELQSL | - | 0.8462 | - | - | - | - | - | - |
|
| Atp6v1a (P50516) | Mus musculus | S384 | YLGARLASFYERAGR | - | 0.0676 | - | - | - | - | - | - |
|
| YAP1 (P46937) | Homo sapiens | S94 | RLRKLPDSFFKPPEP | 0.857 | 0.8235 | - | - | - | - | - | - |
|
| Pde4b (Q8VBU5) | Mus musculus | S125 | SFLYRSDSDYDLSPK | - | 0.4292 | - | - | - | - | - | - |
|
| MAPT (P10636-8) | Homo sapiens | S214 | GSRSRTPSLPTPPTR | 0.068 | 0.9756 | - | - | - | - | - | - |
|
| Ulk1 (O70405) | Mus musculus | S317 | SHLASPPSLGEMPQL | - | 0.7629 | - | - | - | - | - | - |
|
| MAPT (P10636-8) | Homo sapiens | S356 | RVQSKIGSLDNITHV | 0.096 | 0.4831 | - | - | - | - | - | - |
|
| GLI1 (P08151) | Homo sapiens | S408 | GPLPRAPSISTVEPK | 0.213 | 0.8792 | - | - | - | - | - | - |
|
| DDIT3 (P35638) | Homo sapiens | S30 | EDLQEVLSSDENGGT | 0.156 | 0.5707 | - | - | - | - | - | - |
|
| PRPS2 (P11908) | Homo sapiens | S180 | GGAKRVTSIADRLNV | 0.412 | 0.2575 | - | - | - | - | - | - |
|
| PPP1R12C (Q9BZL4) | Homo sapiens | S452 | AGLQRSASSSWLEGT | 0.966 | 0.8623 | - | - | - | - | - | - |
|
| Med1 (Q925J9) | Mus musculus | S671 | SPLERQNSSSGSPRM | - | 0.7951 | - | - | - | - | - | - |
|
| MAPT (P10636-8) | Homo sapiens | S396 | GAEIVYKSPVVSGDT | 0.723 | 0.5992 | - | - | - | - | - | - |
|
| Pde4b (Q8VBU5) | Mus musculus | S118 | GHSQRRESFLYRSDS | - | 0.4441 | - | - | - | - | - | - |
|
| PRPS1 (P60891) | Homo sapiens | S180 | GGAKRVTSIADRLNV | 0.411 | 0.2503 | - | - | - | - | - | - |
|
| TNNI3 (P19429) | Homo sapiens | S23 | PAPIRRRSSNYRAYA | 0.104 | 0.6906 | - | - | - | - | - | - |
|
| Med1 (Q925J9) | Mus musculus | S771 | QRMVRLSSSDSIGPD | - | 0.7629 | - | - | - | - | - | - |
|
| PIAS1 (O75925) | Homo sapiens | S510 | SPVSRTPSLPAVDTS | 0.625 | 0.5665 | - | - | - | - | - | - |
|
| MAPT (P10636-8) | Homo sapiens | S262 | NVKSKIGSTENLKHQ | 0.606 | 0.8125 | - | - | - | - | - | - |
|
| USP10 (Q14694) | Homo sapiens | S76 | DTLPRTPSYSISSTL | 0.045 | 0.7209 | - | - | - | - | - | - |
|
| MKNK1 (Q9BUB5-2) | Homo sapiens | S353 | QVLQRNSSTMDLTLF | 0.097 | 0.4801 | - | - | - | - | - | - |
|
| Braf (P28028) | Mus musculus | S766 | PKIHRSASEPSLNRA | - | 0.4051 | - | - | - | - | - | - |
|
| FANCA (O15360) | Homo sapiens | S347 | VQMQREWSFARTHPL | 0.688 | 0.1914 | - | - | - | - | - | - |
|
| KIF4A (O95239) | Homo sapiens | S801 | KLRRRTFSLTEVRGQ | 0.089 | 0.5951 | - | - | - | - | - | - |
|
| Ulk1 (O70405) | Mus musculus | S777 | TRMFSVGSSSSLGST | - | 0.5992 | - | - | - | - | - | - |
|
| TP73 (O15350) | Homo sapiens | S426 | GGMNKLPSVNQLVGQ | 0.696 | 0.7036 | - | - | - | - | - | - |
|
| Pnpla2 (Q8BJ56) | Mus musculus | S406 | VELRRAQSLPSVPLS | - | 0.3667 | - | - | - | - | - | - |
|
| Gatad2a (Q8CHY6) | Mus musculus | S96 | KSEKRPPSPDVIVLS | - | 0.8421 | - | - | - | - | - | - |
|
| SREBF1 (P36956-3) | Homo sapiens | S372 | TAVHKSKSLKDLVSA | 0.093 | 0.3774 | - | - | - | - | - | - |
|
| TXNIP (Q9H3M7) | Homo sapiens | S308 | GLSSRTSSMASRTSS | 0.392 | 0.6322 | - | - | - | - | - | - |
|
| CGN (Q9P2M7) | Homo sapiens | S131 | GKLLRSHSQASLAGP | 0.977 | 0.5254 | - | - | - | - | - | - |
|
| POU2F1 (P14859) | Homo sapiens | S335 | RFEALNLSFKNMCKL | 0.563 | 0.2064 | - | - | - | - | - | - |
|
| Med1 (Q925J9) | Mus musculus | S811 | GTPVRDSSSSGHSQS | - | 0.7505 | - | - | - | - | - | - |
|
| BRAF (P15056) | Homo sapiens | S729 | PKIHRSASEPSLNRA | 0.947 | 0.4149 | - | - | - | - | - | - |
|
| MAPT (P10636-8) | Homo sapiens | S422 | GSIDMVDSPQLATLA | 0.932 | 0.5211 | - | - | - | - | - | - |
|
| CFTR (P13569) | Homo sapiens | S813 | DIYSRRLSQETGLEI | 0.958 | 0.3356 | - | - | - | - | - | - |
|
| RPTOR (Q8N122) | Homo sapiens | S722 | PRLRSVSSYGNIRAV | 0.794 | 0.3149 | - | - | - | - | - | - |
|
| SYK (P43405) | Homo sapiens | S178 | GKISREESEQIVLIG | 0.348 | 0.2436 | - | - | - | - | - | - |
|
| ACSS2 (Q9NR19) | Homo sapiens | S659 | PGLPKTRSGKIMRRV | 0.849 | 0.494 | - | - | - | - | - | - |
|
| TNNI3 (P19429) | Homo sapiens | S24 | APIRRRSSNYRAYAT | 0.016 | 0.7163 | - | - | - | - | - | - |
|
| JAK1 (P23458) | Homo sapiens | S515 | SLHGSDRSFPSLGDL | 0.235 | 0.3494 | - | - | - | - | - | - |
|
| Nr2c2 (P49117) | Mus musculus | S351 | HVISRDQSTPIIEVE | - | 0.3529 | - | - | - | - | - | - |
|
| SQSTM1 (Q13501) | Homo sapiens | S294 | SSCCSDPSKPGGNVE | 0.532 | 0.8596 | - | - | - | - | - | - |
|
| Map4 (P27546) | Mus musculus | S914 | SVRSKVGSTENIKHQ | - | 0.7982 | - | - | - | - | - | - |
|
| MAPT (P10636-8) | Homo sapiens | T231 | KKVAVVRTPPKSPSS | 0.622 | 0.8966 | - | - | - | - | - | - |
|
| RACK1 (P63244) | Homo sapiens | T50 | WKLTRDETNYGIPQR | 0.009 | 0.2002 | - | - | - | - | - | - |
|
| PAQR3 (Q6TCH7) | Homo sapiens | T32 | PRGIRLYTYEQIPGS | 0.186 | 0.0258 | - | - | - | - | - | - |
|
| EGFR (P00533) | Homo sapiens | T892 | SILHRIYTHQSDVWS | 0.497 | 0.0454 | - | - | - | - | - | - |
|
| ERBB2 (P04626) | Homo sapiens | T900 | SILRRRFTHQSDVWS | 0.432 | 0.078 | - | - | - | - | - | - |
|
| BECN1 (Q14457) | Homo sapiens | T388 | EEVEKGETRFCLPYR | 0.131 | 0.1554 | - | - | - | - | - | - |
|
| Cnbp (P53996) | Mus musculus | T173 | GHLARECTIEATA__ | - | 0.1702 | - | - | - | - | - | - |
|
| EZH2 (Q15910) | Homo sapiens | T311 | NTYKRKNTETALDNK | 0.064 | 0.6755 | - | - | - | - | - | - |
|
| SIRT2 (Q8IXJ6) | Homo sapiens | T101 | PDFRSPSTGLYDNLE | 0.283 | 0.3215 | - | - | - | - | - | - |
|
| CTTN (Q14247) | Homo sapiens | T401 | QARAKTQTPPVSPAP | 0.171 | 0.9126 | - | - | - | - | - | - |
|
| PRKAB1 (Q9Y478) | Homo sapiens | T148 | IVTSQLGTVNNIIQV | 0.405 | 0.374 | - | - | - | - | - | - |
|
| FOXO1 (Q12778) | Homo sapiens | T649 | PHSVKTTTHSWVSG_ | 0.611 | 0.4476 | - | - | - | - | - | - |
|
| CYCS (P62894) | Bos taurus | T29 | EKGGKHKTGPNLHGL | - | 0.5758 | - | - | - | - | - | - |
|
| CNBP (P62633-4) | Homo sapiens | T173 | GHLARECTIEATA__ | 0.221 | 0.1702 | - | - | - | - | - | - | - |
| NFE2L2 (Q16236-2) | Homo sapiens | S542 | HLLKKQLSTLYLEVF | 0.706 | 0.1092 | - | - | - | - | - | - | - |
| MYLK (Q15746-7) | Homo sapiens | S838 | RAIGRLSSMAMISGL | 0.291 | 0.3286 | - | - | - | - | - | - | - |
| (B2RC52) | Homo sapiens | S385 | AKPKFSISPDSLS__ | - | 0.3599 | - | - | - | - | - | - | - |
| (A8K0H7) | Homo sapiens | S715 | FMSSGRQSVLVKSNE | - | 0.1643 | - | - | - | - | - | - | - |
| EEF2K (O00418) | Homo sapiens | S366 | SPQVRTLSGSRPPLL | 0.742 | 0.6227 | - | - | - | - | - | - | - |
| EEF2K (O00418) | Homo sapiens | S78 | SSGSPANSFHFKEAW | 0.374 | 0.3667 | - | - | - | - | - | - | - |
| ULK1 (O75385) | Homo sapiens | S317 | SHLASPPSLGEMQQL | 0.841 | 0.7982 | - | - | - | - | - | - | - |
| RAF1 (P04049) | Homo sapiens | S621 | PKINRSASEPSLHRA | 0.972 | 0.494 | - | - | - | - | - | - | - |
| MAPT (P10636) | Homo sapiens | S255 | TAAREATSIPGFPAE | 0.074 | 0.9081 | - | - | - | - | - | - | - |
| MAPT (P10636) | Homo sapiens | S355 | EADLPEPSEKQPAAA | 0.268 | 0.9519 | - | - | - | - | - | - | - |
| MAPT (P10636) | Homo sapiens | S396 | DDKKAKTSTRSSAKT | 0.723 | 0.8853 | - | - | - | - | - | - | - |
| POU2F1 (P14859) | Homo sapiens | S385 | RRRKKRTSIETNIRV | 0.674 | 0.4369 | - | - | - | - | - | - | - |
| GYS2 (P54840) | Homo sapiens | S8 | MLRGRSLSVTSLGGL | 0.405 | 0.1702 | - | - | - | - | - | - | - |
| ACACA (Q13085-2) | Homo sapiens | S22 | MLQRSSMSGLHLVKQ | 0.000 | 0.2715 | - | - | - | - | - | - | - |
| ACACA (Q13085-4) | Homo sapiens | S117 | LHIRSSMSGLHLVKQ | 0.302 | 0.2541 | - | - | - | - | - | - | - |
| ACACA (Q13085-4) | Homo sapiens | S1238 | IPTLNRMSFSSNLNH | 0.174 | 0.2783 | - | - | - | - | - | - | - |
| SLC12A1 (Q13621) | Homo sapiens | S122 | YRNTGSISGPKVNRP | 0.201 | 0.3215 | - | - | - | - | - | - | - |
| KLC2 (Q9H0B6) | Homo sapiens | S539 | WNGDGSGSLRRSGSF | 0.153 | 0.5707 | - | - | - | - | - | - | - |
| KLC2 (Q9H0B6) | Homo sapiens | S581 | NPRMKRASSLNFLNK | 0.570 | 0.7982 | - | - | - | - | - | - | - |
| ULK1 (O75385) | Homo sapiens | S467 | SAIRRSGSTSPLGFA | 0.864 | 0.7755 | - | - | - | - | - | - | - |
| ULK1 (O75385) | Homo sapiens | S556 | GLGCRLHSAPNLSDL | 0.945 | 0.5665 | - | - | - | - | - | - | - |
| GABBR2 (H9NIL8) | Homo sapiens | S784 | VTSVNQASTSRLEGL | - | 0.5055 | - | - | - | - | - | - | - |
| RB1 (A0A024RDV3) | Homo sapiens | S811 | IYISPLKSPYKISEG | - | 0.3215 | - | - | - | - | - | - | - |
| (A8K8W7) | Homo sapiens | S855 | EPMRRSVSEAALAQP | - | 0.7415 | - | - | - | - | - | - | - |
| KCNB1 (Q14721) | Homo sapiens | S444 | ERAKRNGSIVSMNMK | 0.881 | 0.3774 | - | - | - | - | - | - | - |
| KCNB1 (Q14721) | Homo sapiens | S541 | NKMAKTQSQPILNTK | 0.974 | 0.6755 | - | - | - | - | - | - | - |
| NOS3 (A0A090N8H8) | Homo sapiens | S1177 | TSRIRTQSFSLQERQ | - | 0.433 | - | - | - | - | - | - | - |
| TSC2 (A0A1V1G0E9) | Homo sapiens | S1387 | QPLSKSSSSPELQTL | - | 0.8462 | - | - | - | - | - | - | - |
| TSC2 (X5D2U8) | Homo sapiens | S1321 | QPLSKSSSSPELQTL | - | 0.8655 | - | - | - | - | - | - | - |
| CRY1 (A2I2P0) | Homo sapiens | S71 | ANLRKLNSRLFVIRG | - | 0.1162 | - | - | - | - | - | - | - |
| KCNMA1 (Q5SVJ8) | Homo sapiens | S722 | GRSERDCSCMSGRVR | - | 0.1731 | - | - | - | - | - | - | - |
| MLXIPL (Q9NP71) | Homo sapiens | S556 | TLLRSPGSPQETVPE | 0.881 | 0.725 | - | - | - | - | - | - | - |
| GABBR1 (Q9UBS5) | Homo sapiens | S918 | ELRHQLQSRQQLRSR | 0.073 | 0.6322 | - | - | - | - | - | - | - |
| ACACA (Q13085) | Homo sapiens | S1201 | IPTLNRMSFSSNLNH | 0.142 | 0.2783 | - | - | - | - | - | - | - |
| (Q59GM8) | Homo sapiens | S309 | KKLMHSSSLNNTSIS | - | 0.3529 | - | - | - | - | - | - | - |
| SQSTM1 (Q13501) | Homo sapiens | S349 | SSKEVDPSTGELQSL | 0.783 | 0.8343 | - | - | - | - | - | - | - |
| ATP6V1A (P38606) | Homo sapiens | S384 | YLGARLASFYERAGR | 0.453 | 0.0676 | - | - | - | - | - | - | - |
| (Q59GM8) | Homo sapiens | S130 | SFLYRSDSDYDLSPK | - | 0.4476 | - | - | - | - | - | - | - |
| MED1 (Q15648) | Homo sapiens | S671 | SPLERQNSSSGSPRM | 0.863 | 0.7505 | - | - | - | - | - | - | - |
| (Q59GM8) | Homo sapiens | S123 | GHSQRRESFLYRSDS | - | 0.4864 | - | - | - | - | - | - | - |
| MED1 (Q15648) | Homo sapiens | S771 | QRMVRLSSSDSIGPD | 0.860 | 0.7505 | - | - | - | - | - | - | - |
| BRAF (H7C560) | Homo sapiens | S337 | ILTSPSPSKSIPIPQ | - | 0.6576 | - | - | - | - | - | - | - |
| PNPLA2 (A0A0K0L4Y8) | Homo sapiens | S404 | VELRRVQSLPSVPLS | - | 0.3321 | - | - | - | - | - | - | - |
| GATAD2A (Q86YP4) | Homo sapiens | S100 | KSERRPPSPDVIVLS | 0.536 | 0.8565 | - | - | - | - | - | - | - |
| MED1 (Q15648) | Homo sapiens | S811 | GTPLRDSSSSGHSQS | 0.880 | 0.6851 | - | - | - | - | - | - | - |
| NR2C2 (P49116) | Homo sapiens | S351 | HVISRDQSTPIIEVE | 0.436 | 0.3566 | - | - | - | - | - | - | - |
| MAP4 (Q86Y04) | Homo sapiens | S646 | NVRSKVGSTENIKHQ | - | 0.8085 | - | - | - | - | - | - | - |
| TSC2 (P49815-2) | Homo sapiens | T1228 | PPLPRSNTVASFSSL | 0.909 | 0.5098 | - | - | - | - | - | - | - |
| PRKCQ (Q04759) | Homo sapiens | T538 | LGDAKTNTFCGTPDY | 0.670 | 0.2541 | - | - | - | - | - | - | - |
| MTOR (P42345) | Homo sapiens | T2446 | NKRSRTRTDSYSAGQ | 0.191 | 0.662 | - | - | - | - | - | - | - |
| CYCS (G4XXL9) | Homo sapiens | T29 | EKGGKHKTGPNLHGL | - | 0.5342 | - | - | - | - | - | - | - |
Help Instructions
- Hovering on the "*" in "Substrate" column will display the text if there is a note by the annotator for the given site.
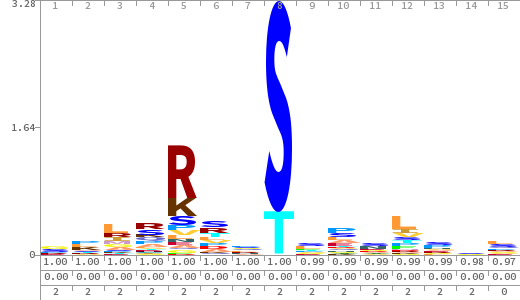
- The column having "Sequence" motif represents phosphosite and its neighbouring 7 residues (upstream and downstream).
- The score in "Conservation" column varies from 0 to 1; Higher the score more conserved is the site.
- The score in "Disorder" column varies from 0 to 1; Score higher than 0.4 suggests disorder.
- In the "Curator Assessment" column, TP refers to True Positive; FP refers to False Positive; and U refers to unknown if the annoatator cannot decide the status based on available information.
- Hovering on the number in the "Evidence Logic" column will show the name of method used to characterize/infer the phosphosite (for more details plase follow the link)