Help Instruction
- Double clicking on the substrate will redirect to the uniprot entry.
- Clicking in the block having the substrate will zoom into its phophosites
- Clicking the center of the circle will lead to zooming out.
- Clicking on the phosphosite will render the phosphosite in its 3D structure (if structure information/mapping is available).
- The color of every substrate has ben kept different in the Sunburst diagram for visual clarity.
- Clicking in the top right corner on the camara icon will download the image.
| Substrate | Organism | PSite | Sequence(+/-7) | Conservation | Disorder | Curator Assessment | Reliability | Evidence Class | Evidence Logic | PubMed | Phospho-ELM | PhosphoSite-Plus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CDC25A (P30304) | Homo sapiens | T507 | KFRTKSRTWAGEKSK | 0.183 | 0.3948 | TP | certain | experimental | 72 | 73 | 140 | 55 | 8 | | 14559997 | - |
|
| CSNK2B (Q5SRQ6) | Homo sapiens | T232 | NFKSPVKTIR_____ | - | 0.3249 | TP | certain | experimental | 73 | 214 | 62 | 74 | | 15225637 | - |
|
| TP53 * (P04637 ) | Homo sapiens | S37 | NVLSPLPSQAMDDLM | 0.107 | 0.708 | U | likely | experimental | 73 | 55 | | 10673501 | - |
|
| (P79923) | Xenopus laevis | S120 | IPIGRRNSLPQNLLG | - | 0.6089 | TP | certain | experimental | 73 | 8 | | 15272308 | - | - |
| (P79923) | Xenopus laevis | S137 | PAFKRNLSYSLECEV | - | 0.2436 | TP | certain | experimental | 73 | 8 | | 15272308 | - | - |
| (P79923) | Xenopus laevis | S190 | GLVQRQKSAPACMLS | - | 0.5665 | TP | certain | experimental | 73 | 8 | | 15272308 | - | - |
| (P79923) | Xenopus laevis | S295 | EKSKRRKSMSEIVNE | - | 0.6375 | TP | certain | experimental | 73 | 8 | | 15272308 | - | - |
| Bad (Q61337) | Mus musculus | S170 | KGLPRPKSAGTATQM | - | 0.8235 | TP | certain | experimental | 73 | 8 | | 15736430 | - | - |
| PPP2R5D (Q14738) | Homo sapiens | S60 | PSSNKRPSNSTPPPT | 0.240 | 0.9649 | TP | certain | experimental | 73 | 8 | 140 | 148 | 55 | 72 | | 17110335 | - |
|
| (P79923) | Xenopus laevis | T504 | KFRMKSRTWAGERSK | - | 0.3117 | TP | certain | experimental | 73 | 8 | 55 | 72 | | 15272308 | - | - |
| Bad (Q61337) | Mus musculus | S155 | GRELRRMSDEFEGSF | - | 0.5456 | TP | certain | experimental | 73 | 8 | 72 | 55 | | 15736430 | - | - |
| TLK1 (Q9UKI8) | Homo sapiens | S743 | PHMRRSNSSGNLHMA | 0.444 | 0.3286 | TP | certain | experimental | 72 | 73 | 9 | 135 | 107 | | 12660173 | - | - |
| CHEK1 * (O14757 ) | Homo sapiens | S296 | GFSKHIQSNLDFSPV | 0.732 | 0.433 | TP | certain | experimental | 72 | 9 | 148 | | 23068608 | - | - |
| CDC25A (P30304) | Homo sapiens | S76 | SNLQRMGSSESTDSG | 0.668 | 0.5707 | TP | certain | experimental | 72 | 107 | 73 | 9 | | 12759351 | - | - |
| CDC25A (P30304) | Homo sapiens | S124 | PALKRSHSDSLDHDI | 0.462 | 0.4476 | TP | certain | experimental | 72 | 107 | 9 | | 12759351 | - | - |
| CDC25A (P30304) | Homo sapiens | S178 | LFTQRQNSAPARMLS | 0.959 | 0.6755 | TP | certain | experimental | 72 | 107 | 9 | | 12759351 | - | - |
| ERRFI1 (Q9UJM3) | Homo sapiens | S251 | RRLRRSHSGPAGSFN | 0.876 | 0.7755 | TP | certain | experimental | 148 | 72 | 73 | 9 | 31 | | 22505024 | - | - |
| ERRFI1 (Q9UJM3) | Homo sapiens | S302 | KPDYRRWSAEVTSST | 0.751 | 0.5901 | TP | certain | experimental | 72 | 31 | | 22505024 | - | - |
| FANCD2 (Q9BXW9) | Homo sapiens | S331 | KSKGRASSSGNQESS | 0.394 | 0.5296 | TP | certain | experimental | 72 | 73 | 148 | 30 | 8 | | 19861535 | - | - |
| NEK11 (Q8NG66) | Homo sapiens | S273 | SMLNKNPSLRPSAIE | 0.578 | 0.2918 | TP | certain | experimental | 72 | 73 | 9 | 55 | 148 | | 19734889 | - | - |
| TLK1 (Q9UKI8-5) | Homo sapiens | S695 | PHMRRSNSSGNLHMA | 0.598 | 0.3286 | TP | certain | experimental | 72 | 73 | 135 | 8 | | 12660173 | - | - |
| E2F6 * (O75461 ) | Homo sapiens | S12 | RPARKLPSLLLDPTE | 0.186 | 0.494 | TP | certain | experimental | 72 | 9 | 73 | 148 | | 23954429 | - | - |
| E2F6 * (O75461 ) | Homo sapiens | S52 | EDNVQYVSMRKALKV | 0.324 | 0.2752 | TP | likely | experimental | 72 | 9 | | 23954429 | - | - |
| RASSF1 (Q9NS23-2) | Homo sapiens | S184 | PSSKKPPSLQDARRG | 0.194 | 0.6948 | TP | certain | experimental | 72 | 148 | 147 | 23 | 30 | 73 | 9 | 143 | | 24197116 | - | - |
| FANCE (Q9HB96) | Homo sapiens | T346 | LGLLRLCTWLLALSP | 0.171 | 0.0269 | TP | certain | experimental | 72 | 9 | 73 | 55 | | 17296736 | - | - |
| E2F3 (O00716-1) | Homo sapiens | S124 | PALGRGGSGGGGGPP | 0.130 | 0.9106 | TP | certain | experimental | 72 | 9 | 73 | 55 | | 19917728 | - | - |
| CDC25A (P30304) | Homo sapiens | S279 | VLKRPERSQEESPPG | 0.549 | 0.8872 | - | - | - | - | - | Non high throughput (12676583) |
|
| CDC25A (P30304) | Homo sapiens | S293 | GSTKRRKSMSGASPK | 0.631 | 0.8713 | - | - | - | - | - | Non high throughput (12676583) |
|
| MCM4 (P33991) | Homo sapiens | S54 | ELQPMPTSPGVDLQS | 0.981 | 0.9211 | - | - | - | - | - | Non high throughput (12714602) | - |
| TP73 (O15350) | Homo sapiens | S47 | EVVGGTDSSMDVFHL | 0.047 | 0.5901 | - | - | - | - | - | Non high throughput (14585975) |
|
| CDC25B (P30305) | Homo sapiens | S230 | AFAQRPSSAPDLMCL | 0.967 | 0.5456 | - | - | - | - | - | Non high throughput (17003105) |
|
| TP53 (P04637) | Homo sapiens | S20 | PLSQETFSDLWKLLP | 0.567 | 0.6183 | - | - | - | - | - | Non high throughput (17339337) |
|
| E2F3 (O00716) | Homo sapiens | S124 | PALGRGGSGGGGGPP | 0.130 | 0.9106 | - | - | - | - | - | - |
|
| MDM4 (O15151) | Homo sapiens | S342 | SKLTHSLSTSDITAI | 0.727 | 0.4292 | - | - | - | - | - | - |
|
| MDM4 (O15151) | Homo sapiens | S367 | PDCRRTISAPVVRPK | 0.922 | 0.433 | - | - | - | - | - | - |
|
| Mdm4 (O35618) | Mus musculus | S367 | PDCRRTISAPVVRPK | - | 0.3872 | - | - | - | - | - | - |
|
| TP53 (P04637) | Homo sapiens | S15 | PSVEPPLSQETFSDL | 0.830 | 0.708 | - | - | - | - | - | - |
|
| TP53 (P04637) | Homo sapiens | S378 | SKKGQSTSRHKKLMF | 0.826 | 0.7881 | - | - | - | - | - | - |
|
| RB1 (P06400) | Homo sapiens | S612 | MYLSPVRSPKKKGST | 0.442 | 0.433 | - | - | - | - | - | - |
|
| MAPT (P10636-8) | Homo sapiens | S262 | NVKSKIGSTENLKHQ | 0.606 | 0.8125 | - | - | - | - | - | - |
|
| NFKB1 (P19838) | Homo sapiens | S328 | INITKPASVFVQLRR | 0.446 | 0.3149 | - | - | - | - | - | - |
|
| CDC25B (P30305) | Homo sapiens | S563 | TFRLKTRSWAGERSR | 0.063 | 0.2715 | - | - | - | - | - | - |
|
| CDC25C (P30307) | Homo sapiens | S216 | SGLYRSPSMPENLNR | 0.868 | 0.6136 | - | - | - | - | - | - |
|
| SYK (P43405) | Homo sapiens | S295 | GIISRIKSYSFPKPG | 0.834 | 0.3983 | - | - | - | - | - | - |
|
| BLM (P54132) | Homo sapiens | S646 | LKHERFQSLSFPHTK | 0.099 | 0.422 | - | - | - | - | - | - |
|
| AURKB (Q96GD4) | Homo sapiens | S331 | HPWVRANSRRVLPPS | 0.298 | 0.3215 | - | - | - | - | - | - |
|
| FANCE (Q9HB96) | Homo sapiens | S374 | LFLGRILSLTSSASR | 0.764 | 0.0543 | - | - | - | - | - | - |
|
| LATS2 (Q9NRM7) | Homo sapiens | S408 | PVPSRTNSFNSHQPR | 0.893 | 0.9013 | - | - | - | - | - | - |
|
| ING1 (Q9UK53-2) | Homo sapiens | S126 | LGDTAGNSGKAGADR | 0.000 | 0.853 | - | - | - | - | - | - |
|
| TLK1 (Q9UKI8-2) | Homo sapiens | S764 | PHMRRSNSSGNLHMA | 0.146 | 0.3286 | - | - | - | - | - | - |
|
| TP53 (P04637) | Homo sapiens | T18 | EPPLSQETFSDLWKL | 0.691 | 0.6183 | - | - | - | - | - | - |
|
| TP53 (P04637) | Homo sapiens | T387 | HKKLMFKTEGPDSD_ | 0.025 | 0.8313 | - | - | - | - | - | - |
|
| CSNK2B (P67870) | Homo sapiens | T213 | NFKSPVKTIR_____ | 0.075 | 0.3249 | - | - | - | - | - | - | - |
| RELA (Q04206) | Homo sapiens | T505 | EAITRLVTGAQRPPD | 0.384 | 0.7951 | - | - | - | - | - | - | - |
| CLSPN (Q9HAW4) | Homo sapiens | T916 | DELLDLCTGKFTSQA | 0.136 | 0.4619 | - | - | - | - | - | - |
|
| Hist1h3a (P68433) | Mus musculus | S11 | TKQTARKSTGGKAPR | - | 0.7505 | - | - | - | - | - | - |
|
| H3F3A (P84243) | Homo sapiens | S32 | AARKSAPSTGGVKKP | 0.000 | 0.6948 | - | - | - | - | - | - |
|
| CTR9 (Q6PD62) | Homo sapiens | S1081 | RRPRRQRSDQDSDSD | 0.373 | 0.986 | - | - | - | - | - | - |
|
| DDX19A (Q9NUU7) | Homo sapiens | S92 | SPLYSVKSFEELRLK | 0.446 | 0.4119 | - | - | - | - | - | - |
|
| BAZ1A (Q9NRL2) | Homo sapiens | S1320 | RSLRKINSAPPTETK | 0.969 | 0.8279 | - | - | - | - | - | - |
|
| BLM (P54132) | Homo sapiens | S539 | DQNKHTASINDLERE | 0.383 | 0.7369 | - | - | - | - | - | - |
|
| TRIP12 (Q14669) | Homo sapiens | S1078 | SGLARAASKDTISNN | 0.405 | 0.6661 | - | - | - | - | - | - |
|
| CLK3 (P49761) | Homo sapiens | S157 | HHCKRYRSPEPDPYL | 0.999 | 0.562 | - | - | - | - | - | - |
|
| EPRS (P07814) | Homo sapiens | S910 | VLFDKVASQGEVVRK | 0.163 | 0.4149 | - | - | - | - | - | - |
|
| SETMAR (Q53H47) | Homo sapiens | S508 | LYDNRRRSAQWLDQE | 0.641 | 0.2988 | - | - | - | - | - | - |
|
| Csnk1d (Q06486) | Rattus norvegicus | S328 | PATRGLPSTASGRLR | - | 0.8085 | - | - | - | - | - | - |
|
| PRR5 (P85299) | Homo sapiens | S240 | RLLRRSRSGDVLAKN | 0.326 | 0.4409 | - | - | - | - | - | - |
|
| GTF3C1 (Q12789) | Homo sapiens | S667 | VRLVRNLSEEGLLRL | 0.671 | 0.2783 | - | - | - | - | - | - |
|
| Cdkn1c (P49919) | Mus musculus | S19 | TAMERLASSDTFPVI | - | 0.422 | - | - | - | - | - | - |
|
| INF2 (Q27J81) | Homo sapiens | S574 | LNWQKLPSNVAREHN | 0.608 | 0.4476 | - | - | - | - | - | - |
|
| TICRR (Q7Z2Z1) | Homo sapiens | S1045 | RSVQRVHSFQQDKSD | 0.886 | 0.8343 | - | - | - | - | - | - |
|
| Csnk1d (Q06486) | Rattus norvegicus | S370 | MERERKVSMRLHRGA | - | 0.7163 | - | - | - | - | - | - |
|
| Csnk1d (Q06486) | Rattus norvegicus | S331 | RGLPSTASGRLRGTQ | - | 0.725 | - | - | - | - | - | - |
|
| TFDP1 (Q14186) | Homo sapiens | S23 | VFIDQNLSPGKGVVS | 0.293 | 0.2988 | - | - | - | - | - | - |
|
| TP53BP1 (Q12888) | Homo sapiens | S1678 | ITSEEERSPAKRGRK | 0.660 | 0.7415 | - | - | - | - | - | - |
|
| ZNF638 (Q14966) | Homo sapiens | S375 | GSKKNYQSQADIPIR | 0.220 | 0.5807 | - | - | - | - | - | - |
|
| SCAF11 (Q99590) | Homo sapiens | S1127 | QQSYKRKSEQEFSFD | 0.044 | 0.5901 | - | - | - | - | - | - |
|
| SRRM2 (Q9UQ35) | Homo sapiens | S2280 | PRTAVAPSAVNLADP | 0.734 | 0.6427 | - | - | - | - | - | - |
|
| GOLGA4 (Q13439) | Homo sapiens | S71 | KLQLRVPSVESLFRS | 0.817 | 0.3426 | - | - | - | - | - | - |
|
| ADD3 (Q9UEY8) | Homo sapiens | S693 | KKKFRTPSFLKKNKK | 0.971 | 0.562 | - | - | - | - | - | - |
|
| KIFC1 (Q9BW19) | Homo sapiens | S26 | RPLIKAPSQLPLSGS | 0.194 | 0.708 | - | - | - | - | - | - |
|
| MATR3 (P43243) | Homo sapiens | S195 | SFDDRGPSLNPVLDY | 0.478 | 0.6136 | - | - | - | - | - | - |
|
| LATS2 (Q9NRM7) | Homo sapiens | S835 | GSHVRQDSMEPSDLW | 0.672 | 0.3426 | - | - | - | - | - | - |
|
| RIF1 (Q5UIP0) | Homo sapiens | S2205 | VNKVRRVSFADPIYQ | 0.812 | 0.3948 | - | - | - | - | - | - |
|
| E2f7 (Q6S7F2) | Mus musculus | S411 | ERLVRYGSFNTVHTS | - | 0.2503 | - | - | - | - | - | - |
|
| CWC25 (Q9NXE8) | Homo sapiens | S337 | PGYTRKLSAEELERK | 0.831 | 0.7881 | - | - | - | - | - | - |
|
| RBM14 (Q96PK6) | Homo sapiens | S627 | SQLSFRRSPTKSSLD | 0.793 | 0.5807 | - | - | - | - | - | - |
|
| IDI1 (Q13907) | Homo sapiens | S55 | GLLHRAFSVFLFNTE | 0.885 | 0.0929 | - | - | - | - | - | - |
|
| CAAP1 (Q9H8G2) | Homo sapiens | S203 | EGDNGMDSDMEEEAD | 0.897 | 0.7951 | - | - | - | - | - | - |
|
| TLK1 (Q9UKI8) | Homo sapiens | S679 | KPFGHNQSQQDILQE | 0.367 | 0.4409 | - | - | - | - | - | - |
|
| PRRC2C (Q9Y520) | Homo sapiens | S661 | QPRPAVLSGYFKQFQ | 0.701 | 0.6322 | - | - | - | - | - | - |
|
| TICRR (Q7Z2Z1) | Homo sapiens | S865 | NALIRHKSIAEVSQN | 0.873 | 0.4801 | - | - | - | - | - | - |
|
| PHF2 (O75151) | Homo sapiens | S929 | REGTRVASIETGLAA | 0.791 | 0.7289 | - | - | - | - | - | - |
|
| HNRNPM (P52272) | Homo sapiens | S432 | HGMDRVGSEIERMGL | 0.975 | 0.422 | - | - | - | - | - | - |
|
| CCDC6 (Q16204) | Homo sapiens | S244 | QPVSAPPSPRDISME | 0.164 | 0.7459 | - | - | - | - | - | - |
|
| WIZ (O95785) | Homo sapiens | S1129 | SLEARSPSDLHISPL | 0.161 | 0.7331 | - | - | - | - | - | - |
|
| NUP153 (P49790) | Homo sapiens | S297 | QMKAKQLSAQSYGVT | 0.029 | 0.5382 | - | - | - | - | - | - |
|
| E2f8 (Q58FA4) | Mus musculus | S395 | PSFTRHPSLIKLVKS | - | 0.4685 | - | - | - | - | - | - |
|
| SLTM (Q9NWH9) | Homo sapiens | S748 | WSENKKLSLDTDARF | 0.035 | 0.5992 | - | - | - | - | - | - |
|
| OGT (O15294) | Homo sapiens | S20 | EPTKRMLSFQGLAEL | 0.202 | 0.4292 | - | - | - | - | - | - |
|
| SSBP1 (Q04837) | Homo sapiens | S67 | ATNEMWRSGDSEVYQ | 0.111 | 0.2436 | - | - | - | - | - | - |
|
| KPNA2 (P52292) | Homo sapiens | S54 | MLKRRNVSSFPDDAT | 0.207 | 0.6322 | - | - | - | - | - | - |
|
| MCM3 (P25205) | Homo sapiens | S160 | KTIERRYSDLTTLVA | 0.013 | 0.2609 | - | - | - | - | - | - |
|
| SRRM2 (Q9UQ35) | Homo sapiens | S1499 | TPRPRSRSPSSPELN | 0.587 | 0.9346 | - | - | - | - | - | - |
|
| CHEK1 (O14757) | Homo sapiens | S316 | EENVKYSSSQPEPRT | 0.572 | 0.6427 | - | - | - | - | - | - |
|
| POP4 (O95707) | Homo sapiens | S10 | SVIYHALSQKEANDS | 0.062 | 0.422 | - | - | - | - | - | - |
|
| ADD2 (P35612) | Homo sapiens | S713 | KKKFRTPSFLKKSKK | 0.935 | 0.5493 | - | - | - | - | - | - |
|
| RAVER1 (Q8IY67-2) | Homo sapiens | S700 | PLGGQKRSFAHLLPS | - | 0.712 | - | - | - | - | - | - |
|
| ADD1 (P35611) | Homo sapiens | S726 | KKKFRTPSFLKKSKK | 0.978 | 0.5992 | - | - | - | - | - | - |
|
| ANKRD12 (Q6UB98-2) | Homo sapiens | S76 | SRNEERDSDTEKEGP | 0.850 | 0.9249 | - | - | - | - | - | - |
|
| ZNF395 (Q9H8N7) | Homo sapiens | S447 | LSPVRSRSLSFSEPQ | 0.708 | 0.4801 | - | - | - | - | - | - |
|
| XRCC6 (P12956) | Homo sapiens | S477 | RFTYRSDSFENPVLQ | 0.233 | 0.2752 | - | - | - | - | - | - |
|
| MAD2L1 (Q13257) | Homo sapiens | S185 | SEEVRLRSFTTTIHK | 0.844 | 0.2951 | - | - | - | - | - | - |
|
| MYCBP2 (O75592) | Homo sapiens | S3478 | TVFQRSYSVVASEYD | 0.481 | 0.3215 | - | - | - | - | - | - |
|
| RPL19 (P84098) | Homo sapiens | S12 | RLQKRLASSVLRCGK | 0.019 | 0.3948 | - | - | - | - | - | - |
|
| BUD13 (Q9BRD0) | Homo sapiens | S18 | EYLKRYLSGADAGVD | 0.861 | 0.4582 | - | - | - | - | - | - |
|
| TBC1D4 (O60343) | Homo sapiens | S588 | RMRGRLGSVDSFERS | 0.583 | 0.6322 | - | - | - | - | - | - |
|
| CCDC6 (Q16204) | Homo sapiens | S240 | EKLDQPVSAPPSPRD | 0.308 | 0.6576 | - | - | - | - | - | - |
|
| DDX24 (Q9GZR7) | Homo sapiens | S82 | KRKAQAVSEEEEEEE | 0.134 | 0.8313 | - | - | - | - | - | - |
|
| CTTN (Q14247) | Homo sapiens | S298 | EKLAKHESQQDYSKG | 0.484 | 0.5456 | - | - | - | - | - | - |
|
| RBM42 (Q9BTD8) | Homo sapiens | S406 | RAFSRFPSFLKAKVI | 0.588 | 0.1399 | - | - | - | - | - | - |
|
| RBM14 (Q96PK6) | Homo sapiens | S618 | LSDYRRLSESQLSFR | 0.161 | 0.4864 | - | - | - | - | - | - |
|
| TRIM28 (Q13263) | Homo sapiens | S473 | SGVKRSRSGEGEVSG | 0.459 | 0.7163 | - | - | - | - | - | - |
|
| CHEK1 (O14757) | Homo sapiens | S291 | SESPSGFSKHIQSNL | 0.426 | 0.4369 | - | - | - | - | - | - |
|
| MYBBP1A (Q9BQG0) | Homo sapiens | S1310 | SLVIRSPSLLQSGAK | 0.000 | 0.725 | - | - | - | - | - | - |
|
| SENP2 (Q9HC62) | Homo sapiens | S344 | GLLRRKVSIIETKEK | 0.945 | 0.391 | - | - | - | - | - | - |
|
| CAMSAP2 (Q08AD1) | Homo sapiens | S464 | RGITRSISNEGLTLN | 0.945 | 0.6851 | - | - | - | - | - | - |
|
| PRRC2B (Q5JSZ5) | Homo sapiens | S166 | RGSSRLLSFSPEEFP | 0.073 | 0.6322 | - | - | - | - | - | - |
|
| LMNA (P02545) | Homo sapiens | S307 | DSLSAQLSQLQKQLA | 0.453 | 0.3948 | - | - | - | - | - | - |
|
| ZBTB11 (O95625) | Homo sapiens | S537 | KRKAVPKSAVQQVAQ | 0.554 | 0.5533 | - | - | - | - | - | - |
|
| PHF8 (Q9UPP1) | Homo sapiens | S904 | REGTRVASIETGLAA | 0.737 | 0.6269 | - | - | - | - | - | - |
|
| RACGAP1 (Q9H0H5) | Homo sapiens | S203 | GPVKKTRSIGSAVDQ | 0.320 | 0.8198 | - | - | - | - | - | - |
|
| DDX19B (Q9UMR2) | Homo sapiens | S93 | SPLYSVKSFEELRLK | 0.424 | 0.4149 | - | - | - | - | - | - |
|
| TLK2 (Q86UE8) | Homo sapiens | S686 | KPFGHNQSQQDILQE | 0.287 | 0.4507 | - | - | - | - | - | - |
|
| ARHGEF2 (Q92974) | Homo sapiens | S172 | LGLRRILSQSTDSLN | 0.967 | 0.4441 | - | - | - | - | - | - |
|
| PPME1 (Q9Y570) | Homo sapiens | S15 | MHLGRLPSRPPLPGS | 0.126 | 0.725 | - | - | - | - | - | - |
|
| MATR3 (P43243) | Homo sapiens | S4 | ____MSKSFQQSSLS | 0.775 | 0.6269 | - | - | - | - | - | - |
|
| PRPF3 (O43395) | Homo sapiens | S161 | EERKKQLSFISPPTP | 0.266 | 0.7799 | - | - | - | - | - | - |
|
| TBC1D4 (O60343) | Homo sapiens | S485 | LVIQRHLSSLTDNEQ | 0.540 | 0.3215 | - | - | - | - | - | - |
|
| PROSER2 (Q86WR7) | Homo sapiens | S43 | SSSSRSRSFTLDDES | 0.178 | 0.5577 | - | - | - | - | - | - |
|
| CDK13 (Q14004) | Homo sapiens | S437 | RSRSRHSSISPSTLT | 0.677 | 0.7755 | - | - | - | - | - | - |
|
| RRP12 (Q5JTH9) | Homo sapiens | S1049 | AKRHRALSQAAVEEE | 0.138 | 0.6227 | - | - | - | - | - | - |
|
| Cdkn1a (P39689) | Mus musculus | S141 | GRKRRQTSLTDFYHS | - | 0.6043 | - | - | - | - | - | - |
|
| DRG2 (P55039) | Homo sapiens | S72 | VALIGFPSVGKSTFL | 0.704 | 0.0765 | - | - | - | - | - | - |
|
| PLRG1 (O43660) | Homo sapiens | S119 | TKIQRMPSESAAQSL | 0.992 | 0.6269 | - | - | - | - | - | - |
|
| HIVEP1 (P15822) | Homo sapiens | S1749 | ASSKRMLSPANSLDI | 0.834 | 0.5951 | - | - | - | - | - | - |
|
| SRRM2 (Q9UQ35) | Homo sapiens | S2388 | TSPRVPLSAYERVSG | 0.345 | 0.6806 | - | - | - | - | - | - |
|
| NCBP3 (Q53F19) | Homo sapiens | S100 | AKRFHFRSEVNLAQR | 0.001 | 0.3983 | - | - | - | - | - | - |
|
| ZBTB11 (O95625) | Homo sapiens | S511 | RSRLRQRSVNEGAYI | 0.865 | 0.4979 | - | - | - | - | - | - |
|
| CDK1 (P06493) | Homo sapiens | S208 | KPLFHGDSEIDQLFR | 0.817 | 0.0985 | - | - | - | - | - | - |
|
| Errfi1 (P05432) | Rattus norvegicus | S250 | RRLRRSHSGPAGSFN | - | 0.7799 | - | - | - | - | - | - |
|
| POLDIP3 (Q9BY77) | Homo sapiens | S63 | ARQKIGLSDARLKLG | 0.065 | 0.3872 | - | - | - | - | - | - |
|
| LIG1 (P18858) | Homo sapiens | S76 | EEEDEALSPAKGQKP | 0.028 | 0.9249 | - | - | - | - | - | - |
|
| KPNA1 (P52294) | Homo sapiens | S18 | LKSYKNKSLNPDEMR | 0.103 | 0.6089 | - | - | - | - | - | - |
|
| SLTM (Q9NWH9) | Homo sapiens | S815 | TARREDPSFERYPKN | 0.037 | 0.7755 | - | - | - | - | - | - |
|
| TICRR (Q7Z2Z1) | Homo sapiens | S1894 | FSRRRPISRTYTRKK | 0.533 | 0.4476 | - | - | - | - | - | - |
|
| OSBPL11 (Q9BXB4) | Homo sapiens | S189 | PISQRRPSQNAISFF | 0.918 | 0.5139 | - | - | - | - | - | - |
|
| UIMC1 (Q96RL1) | Homo sapiens | S627 | HSEGRLLSFLEQSEH | 0.431 | 0.5854 | - | - | - | - | - | - |
|
| SRRM2 (Q9UQ35) | Homo sapiens | S1497 | PQTPRPRSRSPSSPE | 0.366 | 0.9657 | - | - | - | - | - | - |
|
| NUP153 (P49790) | Homo sapiens | S330 | ADAKRIPSIVSSPLN | 0.476 | 0.5176 | - | - | - | - | - | - |
|
| TNK1 (Q13470) | Homo sapiens | S502 | RMKGISRSLESVLSL | 0.996 | 0.6531 | - | - | - | - | - | - |
|
| EML3 (Q32P44) | Homo sapiens | S176 | KLSRKAISSANLLVR | 0.999 | 0.6136 | - | - | - | - | - | - |
|
| CEP170B (Q9Y4F5) | Homo sapiens | S655 | QGLPVPGSPGGQKWV | 0.403 | 0.7289 | - | - | - | - | - | - |
|
| LMO7 (Q8WWI1) | Homo sapiens | S1510 | GIMRRGESLDNLDSP | 0.897 | 0.8085 | - | - | - | - | - | - |
|
| Hist1h3a (P68433) | Mus musculus | T12 | KQTARKSTGGKAPRK | - | 0.7415 | - | - | - | - | - | - |
|
| LAMC1 (P11047) | Homo sapiens | T258 | VTLNRLNTFGDEVFN | 0.818 | 0.3053 | - | - | - | - | - | - |
|
| HNRNPA2B1 (P22626) | Homo sapiens | T159 | KRGFGFVTFDDHDPV | 0.532 | 0.433 | - | - | - | - | - | - |
|
| RPL19 (P84098) | Homo sapiens | T56 | LIIRKPVTVHSRARC | 0.021 | 0.4831 | - | - | - | - | - | - |
|
| YBX1 (P67809) | Homo sapiens | T80 | GFINRNDTKEDVFVH | 0.714 | 0.5807 | - | - | - | - | - | - |
|
| TICRR (Q7Z2Z1) | Homo sapiens | T1898 | RPISRTYTRKKLMGT | 0.204 | 0.422 | - | - | - | - | - | - |
|
| Cdkn1a (P39689) | Mus musculus | T140 | QGRKRRQTSLTDFYH | - | 0.6136 | - | - | - | - | - | - |
|
| RBM26 (Q5T8P6) | Homo sapiens | T839 | TELRRKYTELQLEAA | 0.436 | 0.4619 | - | - | - | - | - | - |
|
| YBX3 (P16989) | Homo sapiens | T112 | GFINRNDTKEDVFVH | 0.864 | 0.5456 | - | - | - | - | - | - |
|
| MKI67 (P46013) | Homo sapiens | T1764 | RSLRKADTEEEFLAF | 0.135 | 0.6661 | - | - | - | - | - | - |
|
| RALY (Q9UKM9) | Homo sapiens | T160 | PLVRRVKTNVPVKLF | 0.082 | 0.3215 | - | - | - | - | - | - |
|
| CHD7 (Q9P2D1) | Homo sapiens | T1227 | AILEKNFTFLSKGGG | 0.003 | 0.1137 | - | - | - | - | - | - |
|
| SEPT7 (Q16181) | Homo sapiens | T228 | KIYEFPETDDEEENK | 0.131 | 0.384 | - | - | - | - | - | - |
|
| SRRM2 (Q9UQ35) | Homo sapiens | T2252 | ARTPAIPTAVNLADS | 0.049 | 0.6269 | - | - | - | - | - | - |
|
| MCM5 (P33992) | Homo sapiens | T633 | MKLQPFATEADVEEA | 0.293 | 0.2258 | - | - | - | - | - | - |
|
| SPEN (Q96T58) | Homo sapiens | T1059 | IKLDRLNTVASPKDC | 0.625 | 0.5382 | - | - | - | - | - | - |
|
| RASSF1 (Q9NS23-2) | Homo sapiens | T38 | ALRIARGTACNPTRQ | 0.276 | 0.3249 | - | - | - | - | - | - |
|
| MED1 (Q15648) | Homo sapiens | T704 | PEKPKHQTEDDFQRE | 0.343 | 0.8623 | - | - | - | - | - | - |
|
| YBX2 (Q9Y2T7) | Homo sapiens | T115 | GFINRNDTKEDVFVH | 0.862 | 0.5493 | - | - | - | - | - | - |
|
| ZC3HC1 (Q86WB0) | Homo sapiens | T84 | AFFSRVETFSSLKWA | 0.302 | 0.1942 | - | - | - | - | - | - |
|
| RASSF1 (Q9NS23-2) | Homo sapiens | T43 | RGTACNPTRQLVPGR | 0.097 | 0.4186 | - | - | - | - | - | - |
|
| Csnk1d (Q06486) | Rattus norvegicus | T397 | QDTSRMSTSQIPGRV | - | 0.9126 | - | - | - | - | - | - |
|
| BLM (P54132) | Homo sapiens | T182 | SHFVRVSTAQKSKKG | 0.470 | 0.708 | - | - | - | - | - | - |
|
| MDC1 (Q14676) | Homo sapiens | T523 | STTVDINTQVEKEVP | 0.587 | 0.7982 | - | - | - | - | - | - |
|
| STK4 (Q13043) | Homo sapiens | T340 | AVGDEMGTVRVASTM | 0.950 | 0.5807 | - | - | - | - | - | - |
|
| MAP2K2 (P36507) | Homo sapiens | T59 | KRLEAFLTQKAKVGE | 0.108 | 0.3704 | - | - | - | - | - | - |
|
| MAD2L1 (Q13257) | Homo sapiens | T187 | EVRLRSFTTTIHKVN | 0.144 | 0.2884 | - | - | - | - | - | - |
|
| FEN1 (P39748) | Homo sapiens | T195 | PVLMRHLTASEAKKL | 0.674 | 0.2918 | - | - | - | - | - | - |
|
| EBNA1BP2 (Q99848) | Homo sapiens | T182 | KYGKKVQTEVLQKRQ | 0.097 | 0.5577 | - | - | - | - | - | - |
|
| RANBP2 (P49792) | Homo sapiens | T1098 | QTIGPRNTFNFGSKN | 0.231 | 0.7331 | - | - | - | - | - | - |
|
| MATR3 (P43243) | Homo sapiens | T150 | LQLKRRRTEEGPTLS | 0.002 | 0.5342 | - | - | - | - | - | - |
|
| CHTOP (Q9Y3Y2) | Homo sapiens | T212 | ALARPVLTKEQLDNQ | 0.392 | 0.5951 | - | - | - | - | - | - |
|
| RBM14 (Q96PK6) | Homo sapiens | T629 | LSFRRSPTKSSLDYR | 0.793 | 0.6183 | - | - | - | - | - | - |
|
| MAP4 (P27816) | Homo sapiens | T925 | PRLSRLATNTSAPDL | 0.739 | 0.7799 | - | - | - | - | - | - |
|
| HDLBP (Q00341) | Homo sapiens | T8 | MSSVAVLTQESFAEH | 0.500 | 0.4541 | - | - | - | - | - | - |
|
| NHS (Q6T4R5) | Homo sapiens | T401 | RKLRRRKTISGIPRR | 0.717 | 0.6991 | - | - | - | - | - | - |
|
| FYTTD1 (Q96QD9) | Homo sapiens | T205 | KVQAQLNTEQLLDDV | 0.072 | 0.3872 | - | - | - | - | - | - |
|
| RPL13 (P26373) | Homo sapiens | T170 | KEKARVITEEEKNFK | 0.790 | 0.3087 | - | - | - | - | - | - |
|
| MINK1 (Q8N4C8) | Homo sapiens | T280 | TYLSRPPTEQLLKFP | 0.926 | 0.247 | - | - | - | - | - | - |
|
| MAP2K1 (Q02750) | Homo sapiens | T55 | KRLEAFLTQKQKVGE | 0.059 | 0.433 | - | - | - | - | - | - |
|
| SIN3A (Q96ST3) | Homo sapiens | T434 | IRRHPTGTTPPVKKK | 0.115 | 0.6661 | - | - | - | - | - | - |
|
| TLK2 (Q86UE8) | Homo sapiens | S750 | PHIRKSVSTSSPAGA | 0.334 | 0.2503 | - | - | - | - | - | - | - |
| LATS2 (Q9NRM7) | Homo sapiens | S172 | TPVTRRPSFEGTGDS | 0.526 | 0.6183 | - | - | - | - | - | - | - |
| LATS2 (Q9NRM7) | Homo sapiens | S380 | ATLARRDSLQKPGLE | 0.706 | 0.7547 | - | - | - | - | - | - | - |
| LATS2 (Q9NRM7) | Homo sapiens | S446 | HILHPVKSVRVLRPE | 0.824 | 0.6183 | - | - | - | - | - | - | - |
| MDM4 (A0A024R990) | Homo sapiens | S367 | PDCRRTISAPVVRPK | - | 0.433 | - | - | - | - | - | - | - |
| CDC25A (P30304) | Homo sapiens | S107 | NPMRRIHSLPQKLLG | 0.691 | 0.5382 | - | - | - | - | - | - | - |
| CDC25A (P30304) | Homo sapiens | S298 | RKSMSGASPKESTNP | 0.371 | 0.8375 | - | - | - | - | - | - | - |
| (B4DZQ9) | Homo sapiens | S118 | GRELRRMSDEFVDSF | - | 0.6427 | - | - | - | - | - | - | - |
| BAD (Q92934) | Homo sapiens | S134 | KGLPRPKSAGTATQM | 0.882 | 0.8279 | - | - | - | - | - | - | - |
| TP53 (K7PPA8) | Homo sapiens | S20 | PLSQETFSDLWKLLP | - | 0.6183 | - | - | - | - | - | - | - |
| TP53 (K7PPA8) | Homo sapiens | S37 | NVLSPLPSQAMDDLM | - | 0.708 | - | - | - | - | - | - | - |
| CDC25A (A0A024R2S4) | Homo sapiens | S178 | LFTQRQNSAPARMLS | - | 0.6755 | - | - | - | - | - | - | - |
| CDC25A (A0A024R2S4) | Homo sapiens | S279 | VLKRPERSQEESPPG | - | 0.8872 | - | - | - | - | - | - | - |
| (A8K8Z3) | Homo sapiens | S743 | PHMRRSNSSGNLHMA | - | 0.3286 | - | - | - | - | - | - | - |
| CSNK1D (H7BYT1) | Homo sapiens | S328 | PATRGLPSTASGRLR | - | 0.8235 | - | - | - | - | - | - | - |
| CSNK1D (P48730) | Homo sapiens | S370 | MERERKVSMRLHRGA | 0.906 | 0.712 | - | - | - | - | - | - | - |
| CSNK1D (H7BYT1) | Homo sapiens | S331 | RGLPSTASGRLRGTQ | - | 0.7415 | - | - | - | - | - | - | - |
| (Q59ED0) | Homo sapiens | S173 | GRKRRQTSMTDFYHS | - | 0.7163 | - | - | - | - | - | - | - |
| ERRFI1 (I6S2Y9) | Homo sapiens | S251 | RRLRRSHSGPAGSFN | - | 0.7755 | - | - | - | - | - | - | - |
| BRCA2 (P51587) | Homo sapiens | T3387 | LRLKRRCTTSLIKEQ | 0.831 | 0.4119 | - | - | - | - | - | - | - |
| HIST1H3A (P68431) | Homo sapiens | T12 | KQTARKSTGGKAPRK | 0.620 | 0.7415 | - | - | - | - | - | - | - |
| (Q5U0A5) | Homo sapiens | T309 | LRKGRGETRICKIYD | - | 0.1323 | - | - | - | - | - | - | - |
| (Q59ED0) | Homo sapiens | T172 | QGRKRRQTSMTDFYH | - | 0.725 | - | - | - | - | - | - | - |
| CSNK1D (P48730) | Homo sapiens | T397 | QDTSRMSTSQIPGRV | 0.516 | 0.9106 | - | - | - | - | - | - | - |
Help Instructions
- Hovering on the "*" in "Substrate" column will display the text if there is a note by the annotator for the given site.
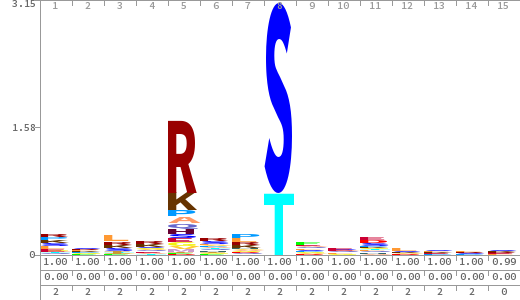
- The column having "Sequence" motif represents phosphosite and its neighbouring 7 residues (upstream and downstream).
- The score in "Conservation" column varies from 0 to 1; Higher the score more conserved is the site.
- The score in "Disorder" column varies from 0 to 1; Score higher than 0.4 suggests disorder.
- In the "Curator Assessment" column, TP refers to True Positive; FP refers to False Positive; and U refers to unknown if the annoatator cannot decide the status based on available information.
- Hovering on the number in the "Evidence Logic" column will show the name of method used to characterize/infer the phosphosite (for more details plase follow the link)