Help Instruction
- Double clicking on the substrate will redirect to the uniprot entry.
- Clicking in the block having the substrate will zoom into its phophosites
- Clicking the center of the circle will lead to zooming out.
- Clicking on the phosphosite will render the phosphosite in its 3D structure (if structure information/mapping is available).
- The color of every substrate has ben kept different in the Sunburst diagram for visual clarity.
- Clicking in the top right corner on the camara icon will download the image.
| Substrate | Organism | PSite | Sequence(+/-7) | Conservation | Disorder | Curator Assessment | Reliability | Evidence Class | Evidence Logic | PubMed | Phospho-ELM | PhosphoSite-Plus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KCND2 (Q9NZV8) | Homo sapiens | S459 | LLSNQLQSSEDEQAF | 0.097 | 0.4369 | TP | certain | experimental | 73 | 273 | 135 | 62 | | 15071113 | - | - |
| Nos1 (Q9Z0J4) | Mus musculus | S847 | SYKVRFNSVSSYSDS | - | 0.3356 | TP | certain | experimental | 72 | 73 | 55 | | 12630910 | - | - |
| PLCB3 * (Q01970 ) | Homo sapiens | S537 | PSLEPQKSLGDEGLN | 0.766 | 0.8623 | TP | certain | experimental | 72 | 107 | 62 | | 11325525 | - | - |
| Nlgn1 (Q99K10) | Mus musculus | T739 | CSPQRTTTNDLTHAP | - | 0.8681 | TP | certain | experimental | 72 | 9 | 31 | 73 | 55 | | 24336150 | - | - |
| TH (P07101) | Homo sapiens | S19 | KGFRRAVSELDAKQA | 0.455 | 0.5665 | TP | certain | experimental | 73 | 139 | | 1680128 | Non high throughput (1680128) |
|
| TH (P07101) | Homo sapiens | S71 | RFIGRRQSLIEDARK | 0.945 | 0.6183 | TP | certain | experimental | 73 | 139 | | 1680128 | Non high throughput (1680128) |
|
| Pln * (P61014 ) | Mus musculus | S16 | RSAIRRASTIEMPQQ | - | 0.087 | U | likely | experimental | 73 | 55 | 9 | | 10988285 | Non high throughput (2544595) | - |
| Gabrg2 (P22723) | Mus musculus | S381 | NPLLRMFSFKAPTID | - | 0.0909 | TP | certain | experimental | 273 | 107 | 9 | | 8027073 | Non high throughput (8027073) | - |
| Gabrg2 (P22723) | Mus musculus | S393 | TIDIRPRSATIQMNN | - | 0.3426 | TP | certain | experimental | 9 | 73 | | 8027073 | Non high throughput (8027073) | - |
| Gabrb1 (P50571) | Mus musculus | S409 | IQYRKPLSSREGFGR | - | 0.2752 | TP | certain | experimental | 73 | 9 | 273 | | 8027073 | Non high throughput (8027073) | - |
| Gabrb1 (P50571) | Mus musculus | S434 | GRIRRRASQLKVKIP | - | 0.4541 | TP | certain | experimental | 73 | 9 | 273 | | 8027073 | Non high throughput (8027073) | - |
| Gabrg2 (P22723) | Mus musculus | T395 | DIRPRSATIQMNNAT | - | 0.3019 | TP | certain | experimental | 9 | 73 | | 8027073 | Non high throughput (8027073) | - |
| ERBB2 (P04626) | Homo sapiens | T1172 | ATLERPKTLSPGKNG | 0.641 | 0.7916 | TP | certain | experimental | 72 | 73 | 8 | 107 | 273 | | 8700533 | Non high throughput (8700533) | - |
| ITPKA (P23677) | Homo sapiens | T311 | EHAQRAVTKPRYMQW | 0.826 | 0.5098 | TP | certain | experimental | 73 | 72 | 148 | 135 | | 9155020 | Non high throughput (9155020) |
|
| EGFR (P00533) | Homo sapiens | S1070 | DSFLQRYSSDPTGAL | 0.500 | 0.5707 | TP | certain | experimental | 73 | 9 | 72 | | 10347170 | Non high throughput (10347170) | - |
| EGFR (P00533) | Homo sapiens | S1071 | SFLQRYSSDPTGALT | 0.500 | 0.6661 | TP | certain | experimental | 73 | 9 | 72 | | 10347170 | Non high throughput (10347170) | - |
| EGFR (P00533) | Homo sapiens | S1081 | TGALTEDSIDDTFLP | 0.500 | 0.712 | TP | certain | experimental | 73 | 9 | 72 | | 10347170 | Non high throughput (10347170) | - |
| EGFR (P00533) | Homo sapiens | S1166 | QKGSHQISLDNPDYQ | 0.452 | 0.5992 | TP | certain | experimental | 73 | 9 | 72 | | 10347170 | Non high throughput (10347170) | - |
| EGFR (P00533) | Homo sapiens | S768 | DEAYVMASVDNPHVC | 0.574 | 0.0704 | TP | certain | experimental | 9 | 73 | | 10347170 | Non high throughput (10347170) | - |
| HSF1 (Q00613) | Homo sapiens | S230 | PKYSRQFSLEHVHGS | 0.584 | 0.5055 | TP | certain | experimental | 72 | 107 | 135 | 73 | 214 | 9 | 55 | | 11447121 | Non high throughput (11447121) |
|
| ALOX5 * (P09917 ) | Homo sapiens | S272 | CSLERQLSLEQEVQQ | 0.144 | 0.1852 | U | likely | experimental | 73 | | 11844797 | Non high throughput (11844797) |
|
| ETS1 (P14921) | Homo sapiens | S251 | GKLGGQDSFESIESY | 0.994 | 0.4017 | TP | certain | experimental | 73 | 9 | 72 | 55 | | 12475968 | Non high throughput (12475968) |
|
| ETS1 (P14921) | Homo sapiens | S257 | DSFESIESYDSCDRL | 0.404 | 0.4292 | TP | certain | experimental | 73 | 9 | 72 | 55 | | 12475968 | Non high throughput (12475968) |
|
| ETS1 (P14921) | Homo sapiens | S282 | NSLQRVPSYDSFDSE | 0.369 | 0.3704 | TP | certain | experimental | 73 | 9 | 72 | 55 | | 12475968 | Non high throughput (12475968) |
|
| ETS1 (P14921) | Homo sapiens | S285 | QRVPSYDSFDSEDYP | 0.381 | 0.3704 | TP | certain | experimental | 73 | 9 | 72 | 55 | | 12475968 | Non high throughput (12475968) |
|
| PPP1R14A * (Q96A00 ) | Homo sapiens | S130 | GLRQPSPSHDGSLSP | 0.144 | 0.8765 | TP | likely | experimental | 73 | 214 | 135 | | 12604330 | Non high throughput (12604330) |
|
| RYR2 (Q92736) | Homo sapiens | S2808 | YNRTRRISQTSQVSV | 0.753 | 0.346 | TP | certain | experimental | 73 | 55 | | 14514795 | Non high throughput (14514795) |
|
| Myog * (P12979 ) | Mus musculus | T87 | VDRRRAATLREKRRL | - | 0.4476 | TP | certain | experimental | 72 | 73 | 55 | | 14966278 | Non high throughput (14751541) | - |
| CLCN3 * (P51790 ) | Homo sapiens | S109 | ERHRRINSKKKESAW | 0.128 | 0.4864 | TP | certain | experimental | 73 | 72 | 9 | | 14754994 | Non high throughput (14754994) |
|
| HRH1 * (P35367 ) | Homo sapiens | S396 | FTWKRLRSHSRQYVS | 0.321 | 0.0719 | TP | likely | experimental | 73 | 9 | | 15107581 | Non high throughput (15107581) |
|
| HRH1 * (P35367 ) | Homo sapiens | S398 | WKRLRSHSRQYVSGL | 0.940 | 0.1643 | TP | likely | experimental | 73 | 9 | | 15107581 | Non high throughput (15107581) |
|
| HRH1 * (P35367 ) | Homo sapiens | T140 | LRYLKYRTKTRASAT | 0.825 | 0.0618 | TP | likely | experimental | 73 | 9 | | 15107581 | Non high throughput (15107581) |
|
| HRH1 * (P35367 ) | Homo sapiens | T142 | YLKYRTKTRASATIL | 0.164 | 0.0223 | TP | likely | experimental | 73 | 9 | | 15107581 | Non high throughput (15107581) |
|
| Il6st * (Q00560 ) | Mus musculus | S780 | QVFSRSESTQPLLDS | - | 0.6661 | TP | certain | experimental | 73 | 148 | 55 | | 16036214 | Non high throughput (16036214) | - |
| CAMK2A * (Q9UQM7 ) | Homo sapiens | T286 | SCMHRQETVDCLKKF | 0.782 | 0.2752 | TP | certain | experimental | 112 | | 16172120 | Non high throughput (16172120) |
|
| Nos1 * (P29476 ) | Rattus norvegicus | S847 | SYKVRFNSVSSYSDS | - | 0.3356 | TP | certain | experimental | 72 | 73 | 55 | 8 | | 10400690 | Non high throughput (10400690, 10874031, 12630910) | - |
| NOS1 (P29475) | Homo sapiens | S852 | SYKVRFNSVSSYSDS | 0.880 | 0.346 | - | - | - | - | 10400690, 10874031, 12630910 | Non high throughput (10400690, 10874031, 12630910) | - |
| Pln * (P61014 ) | Mus musculus | T17 | SAIRRASTIEMPQQA | - | 0.1731 | TP | certain | experimental | 73 | 9 | 148 | | 10988285 | Non high throughput (10988285, 2544595) | - |
| VIM (P08670) | Homo sapiens | S83 | GVRLLQDSVDFSLAD | 0.105 | 0.2849 | TP | certain | experimental | 72 | 55 | 148 | | 16140754 | Non high throughput (16140754, 9794428) |
|
| Ryr2 (B0LPN4) | Rattus norvegicus | S2798 | YNRTRRISQTSQVSI | - | 0.3149 | - | - | - | - | - | - |
|
| Ryr2 (B0LPN4) | Rattus norvegicus | S2804 | ISQTSQVSIDAAHGY | - | 0.4652 | - | - | - | - | - | - |
|
| Ryr2 (E9Q401) | Mus musculus | S2807 | YNRTRRISQTSQVSI | - | 0.3087 | - | - | - | - | - | - |
|
| Trpv1 (O35433) | Rattus norvegicus | S502 | YFLQRRPSLKSLFVD | - | 0.0086 | TP | certain | experimental | 72 | 73 | 8 | | 14630912 | - |
|
| Syn1 (O88935) | Mus musculus | S605 | AGPTRQASQAGPGPR | - | 0.9921 | - | - | - | - | - | - |
|
| NEFL (P02548) | Bos taurus | S58 | LSVRRSYSSSSGSLM | - | 0.391 | TP | certain | experimental | 73 | 214 | 55 | 72 | 148 | | 10854283 | - |
|
| NEFL * (P02548 ) | Bos taurus | S67 | SSGSLMPSLESLDLS | - | 0.3494 | TP | certain | experimental | 73 | 214 | 55 | | 10854283 | - |
|
| NEFL * (P02548 ) | Bos taurus | S70 | SLMPSLESLDLSQVA | - | 0.3215 | TP | certain | experimental | 73 | 214 | 55 | | 10854283 | - |
|
| CALD1 (P12957) | Gallus gallus | S635 | AVVSKIDSRLEQYTS | - | 0.5098 | TP | certain | experimental | 73 | 273 | | 2170388 | - |
|
| TH * (P07101-3 ) | Homo sapiens | S40 | RFIGRRQSLIEDARK | 0.090 | 0.5296 | TP | certain | experimental | 73 | 139 | | 9553069 | - |
|
| CACNA1S * (P07293 ) | Oryctolagus cuniculus | S1575 | PEIRRTISGDLTAEE | - | 0.5493 | TP | certain | experimental | 73 | 214 | | 20937870 | - |
|
| Gja1 * (P08050 ) | Rattus norvegicus | S244 | KDRVKGRSDPYHATT | - | 0.4652 | TP | certain | experimental | 73 | 214 | | 21158428 | - |
|
| Gja1 (P08050) | Rattus norvegicus | S330 | AGSTISNSHAQPFDF | - | 0.7163 | TP | certain | experimental | 73 | 214 | | 21158428 | - |
|
| Gja1 (P08050) | Rattus norvegicus | S364 | AIVDQRPSSRASSRA | - | 0.7982 | TP | certain | experimental | 73 | 214 | | 21158428 | - |
|
| Gja1 (P08050) | Rattus norvegicus | S365 | IVDQRPSSRASSRAS | - | 0.8125 | TP | certain | experimental | 73 | 214 | | 21158428 | - |
|
| Gja1 (P08050) | Rattus norvegicus | S369 | RPSSRASSRASSRPR | - | 0.9362 | TP | certain | experimental | 73 | 214 | | 21158428 | - |
|
| Gja1 (P08050) | Rattus norvegicus | S372 | SRASSRASSRPRPDD | - | 0.8765 | TP | certain | experimental | 73 | 214 | | 21158428 | - |
|
| Gja1 (P08050) | Rattus norvegicus | S373 | RASSRASSRPRPDDL | - | 0.8713 | TP | certain | experimental | 73 | 214 | | 21158428 | - |
|
| Gja1 (P08050) | Rattus norvegicus | S255 | HATTGPLSPSKDCGS | - | 0.4292 | TP | certain | experimental | 73 | 214 | | 21158428 | - |
|
| Gja1 (P08050) | Rattus norvegicus | S257 | TTGPLSPSKDCGSPK | - | 0.2817 | TP | certain | experimental | 73 | 214 | | 21158428 | - |
|
| Gja1 (P08050) | Rattus norvegicus | S296 | VTGDRNNSSCRNYNK | - | 0.4507 | TP | certain | experimental | 73 | 214 | | 21158428 | - |
|
| Gja1 (P08050) | Rattus norvegicus | S297 | TGDRNNSSCRNYNKQ | - | 0.5577 | TP | certain | experimental | 73 | 214 | | 21158428 | - |
|
| Gja1 (P08050) | Rattus norvegicus | S306 | RNYNKQASEQNWANY | - | 0.4441 | TP | certain | experimental | 73 | 214 | | 21158428 | - |
|
| Gja1 (P08050) | Rattus norvegicus | S314 | EQNWANYSAEQNRMG | - | 0.6482 | TP | certain | experimental | 73 | 214 | | 21158428 | - |
|
| Gja1 (P08050) | Rattus norvegicus | S325 | NRMGQAGSTISNSHA | - | 0.7459 | TP | certain | experimental | 73 | 214 | | 21158428 | - |
|
| Gja1 (P08050) | Rattus norvegicus | S328 | GQAGSTISNSHAQPF | - | 0.6991 | TP | certain | experimental | 73 | 214 | | 21158428 | - |
|
| MPZ * (P10522 ) | Bos taurus | S210 | HKTAKDASKRGRQTP | - | 0.4476 | TP | certain | experimental | 73 | 173 | 139 | | 1700069 | - |
|
| MPZ * (P10522 ) | Bos taurus | S233 | SRSTKAASEKKTKGL | - | 0.5665 | TP | certain | experimental | 73 | 173 | 139 | | 1700069 | - |
|
| MAPT * (P10636-8 ) | Homo sapiens | S262 | NVKSKIGSTENLKHQ | 0.606 | 0.8125 | TP | certain | experimental | 73 | 214 | | 12464279 | - |
|
| MAPT * (P10636-8 ) | Homo sapiens | S416 | SNVSSTGSIDMVDSP | 0.459 | 0.5577 | TP | certain | experimental | 72 | 73 | 55 | | 16000144 | - |
|
| Camk2a * (P11275 ) | Rattus norvegicus | S314 | MLATRNFSGGKSGGN | - | 0.5139 | TP | certain | - | 73 | 273 | | 2162839 | - |
|
| Mylk (P11799) | Gallus gallus | S1749 | RAIGRLSSMAMISGM | - | 0.3704 | - | - | - | - | - | - |
|
| Mylk (P11799) | Gallus gallus | S1762 | GMSGRKASGSSPTSP | - | 0.6136 | - | - | - | - | - | - |
|
| SRF (P11831) | Homo sapiens | S103 | RGLKRSLSEMEIGMV | 0.283 | 0.494 | - | - | - | - | - | - |
|
| CALD1 (P12957) | Gallus gallus | S26 | RLEAERLSYQRNDDD | - | 0.6851 | TP | certain | experimental | 73 | 273 | | 2170388 | - |
|
| CALD1 (P12957) | Gallus gallus | S490 | LTTPKLKSTENAFGR | - | 0.7369 | TP | certain | experimental | 73 | 273 | | 2170388 | - |
|
| CALD1 * (P12957 ) | Gallus gallus | S59 | QKEEGDVSGEVTEKS | - | 0.6851 | TP | certain | experimental | 73 | 273 | | 2170388 | - |
|
| CALD1 (P12957) | Gallus gallus | S602 | CFSPKGSSLKIEERA | - | 0.4149 | TP | certain | experimental | 73 | 273 | | 2170388 | - |
|
| CALD1 * (P12957 ) | Gallus gallus | S73 | SEVNAQNSVAEEETK | - | 0.8125 | TP | certain | experimental | 73 | 273 | | 2170388 | - |
|
| CALD1 (P12957) | Gallus gallus | S741 | RNLWEKQSVEKPAAS | - | 0.7459 | TP | certain | experimental | 73 | 273 | | 2170388 | - |
|
| ETS2 (P15036) | Homo sapiens | S246 | FPKSRLSSVSVTYCS | 0.040 | 0.1914 | TP | certain | experimental | 55 | 214 | 154 | 73 | | 19182667 | - |
|
| ETS2 (P15036) | Homo sapiens | S310 | LDVQRVPSFESFEDD | 0.627 | 0.3182 | TP | certain | experimental | 55 | 214 | 154 | 73 | | 19182667 | - |
|
| ETS2 * (P15036 ) | Homo sapiens | S313 | QRVPSFESFEDDCSQ | 0.957 | 0.2436 | TP | certain | experimental | 55 | 214 | 154 | 73 | | 19182667 | - |
|
| Lipe * (P15304 ) | Rattus norvegicus | S865 | ESMRRSVSEAALAQP | - | 0.5807 | TP | certain | experimental | 73 | 8 | | 9636039 | - |
|
| Creb1 (P15337) | Rattus norvegicus | S133 | EILSRRPSYRKILND | - | 0.4979 | TP | certain | experimental | 73 | 8 | | 11013247 | - |
|
| Creb1 * (P15337 ) | Rattus norvegicus | S142 | RKILNDLSSDAPGVP | - | 0.5707 | TP | certain | experimental | 73 | 8 | | 11013247 | - |
|
| CACNA1C (P15381) | Oryctolagus cuniculus | S1517 | DYLTRDWSILGPHHL | - | 0.0631 | - | - | experimental | 73 | 214 | | 16537462 | - |
|
| CACNA1C (P15381) | Oryctolagus cuniculus | S1575 | VACKRLVSMNMPLNS | - | 0.2645 | - | - | - | - | - | - |
|
| CACNA1C (P15381) | Oryctolagus cuniculus | S1700 | PEIRRAISGDLTAEE | - | 0.4901 | TP | certain | experimental | 73 | 214 | 55 | 72 | 8 | | 20876873 | - |
|
| CACNA1C * (P15381 ) | Oryctolagus cuniculus | S439 | GVLSGEFSKEREKAK | - | 0.2918 | TP | certain | experimental | 73 | 214 | | 16537462 | - |
|
| CACNA1C (P15381-4) | Oryctolagus cuniculus | S1512 | DYLTRDWSILGPHHL | - | 0.0631 | TP | certain | experimental | 73 | 55 | 8 | | 16820363 | - |
|
| CACNA1C (P15381-4) | Oryctolagus cuniculus | S1570 | VACKRLVSMNMPLNS | - | 0.2645 | TP | certain | experimental | 73 | 55 | 8 | | 16820363 | - |
|
| CD44 * (P16070-12 ) | Homo sapiens | S325 | LNGEASKSQEMVHLV | 0.089 | 0.6906 | TP | certain | experimental | 72 | 73 | 55 | 8 | 148 | | 11463356 | - |
|
| Ptpra * (P18052 ) | Mus musculus | S180 | QAGSHSNSFRLSNGR | - | 0.5577 | TP | certain | experimental | 73 | 148 | 8 | 72 | 55 | | 18809727 | - |
|
| Ptpra (P18052) | Mus musculus | S204 | PLLARSPSTNRKYPP | - | 0.4619 | TP | certain | experimental | 73 | 148 | 8 | 72 | 55 | | 18809727 | - |
|
| Spr (P18297) | Rattus norvegicus | S196 | EPSVRVLSYAPGPLD | - | 0.3529 | TP | certain | experimental | 73 | 427 | 62 | | 11825621 | - |
|
| Spr (P18297) | Rattus norvegicus | S214 | QQLARETSMDPELRS | - | 0.5211 | TP | certain | experimental | 73 | 427 | 62 | | 11825621 | - |
|
| Spr (P18297) | Rattus norvegicus | S46 | LLSARSDSMLRQLKE | - | 0.1028 | TP | certain | experimental | 73 | 427 | 62 | | 11825621 | - |
|
| Drd3 (P19020) | Rattus norvegicus | S229 | RILTRQNSQCISIRP | - | 0.3019 | TP | certain | experimental | 73 | 72 | 8 | 214 | 23 | 66 | | 19217379 | - |
|
| Gria1 (P19490) | Rattus norvegicus | S645 | LTVERMVSPIESAED | - | 0.1275 | TP | certain | experimental | 73 | 8 | 107 | | 7877986 | - |
|
| Gria1 * (P19490 ) | Rattus norvegicus | S849 | FCLIPQQSINEAIRT | - | 0.1881 | TP | certain | experimental | 73 | 107 | 72 | 55 | 8 | | 9405465 | - |
|
| Gria4 (P19493) | Rattus norvegicus | S862 | TRNKARLSITGSVGE | - | 0.4831 | TP | certain | experimental | 73 | 62 | 107 | | 10366608 | - |
|
| FLNA (P21333) | Homo sapiens | S2523 | VTGPRLVSNHSLHET | 0.139 | 0.5055 | TP | certain | experimental | 72 | 148 | 74 | | 11290523 | - |
|
| Gjc1 * (P28229 ) | Mus musculus | S326 | AQEQQYGSHEEHLPA | - | 0.5665 | TP | certain | experimental | 73 | 214 | | 22127232 | - |
|
| TCAP (O15273) | Homo sapiens | S157 | GALRRSLSRSMSQEA | 0.250 | 0.6806 | - | - | - | - | - | - |
|
| Gjc1 (P28229) | Mus musculus | S381 | SKSGSNKSSISSKSG | - | 0.7672 | TP | certain | experimental | 73 | 214 | | 22127232 | - |
|
| Gjc1 * (P28229 ) | Mus musculus | S382 | KSGSNKSSISSKSGD | - | 0.7672 | TP | certain | experimental | 73 | 214 | | 22127232 | - |
|
| Gjc1 * (P28229 ) | Mus musculus | S384 | GSNKSSISSKSGDGK | - | 0.7672 | TP | certain | experimental | 73 | 214 | | 22127232 | - |
|
| Gjc1 (P28229) | Mus musculus | S385 | SNKSSISSKSGDGKT | - | 0.6576 | TP | certain | experimental | 73 | 214 | | 22127232 | - |
|
| Gjc1 * (P28229 ) | Mus musculus | S387 | KSSISSKSGDGKTSV | - | 0.562 | TP | certain | experimental | 73 | 214 | | 22127232 | - |
|
| Gjc1 * (P28229 ) | Mus musculus | S393 | KSGDGKTSVWI____ | - | 0.422 | TP | certain | experimental | 73 | 214 | | 22127232 | - |
|
| MAPT * (P29172 ) | Bos taurus | S423 | SNVSSTGSIDMVDSP | - | 0.5577 | - | - | - | 73 | 55 | | - | - |
|
| Vim * (P31000 ) | Rattus norvegicus | S39 | TTSTRTYSLGSALRP | - | 0.4685 | TP | certain | experimental | 427 | | 16629905 | - |
|
| Dlg4 (P31016) | Rattus norvegicus | S73 | ITLERGNSGLGFSIA | - | 0.3631 | TP | certain | experimental | 72 | 73 | 8 | 55 | 427 | | 17156196 | - |
|
| Oprm1 (P33535) | Rattus norvegicus | S261 | LMILRLKSVRMLSGS | - | 0.0136 | TP | certain | experimental | 73 | 8 | | 9326307 | - |
|
| Oprm1 (P33535) | Rattus norvegicus | S266 | LKSVRMLSGSKEKDR | - | 0.0909 | TP | certain | experimental | 73 | 8 | | 9326307 | - |
|
| Ptk2 (P34152-3) | Mus musculus | S843 | DVRLSRGSIDREDGS | - | 0.6806 | TP | certain | experimental | 72 | 73 | 8 | 55 | 148 | 140 | | 15845548 | - |
|
| SPR (P35270) | Homo sapiens | S213 | QQLARETSVDPDMRK | 0.339 | 0.5254 | TP | certain | experimental | 73 | 62 | | 11825621 | - |
|
| OPRM1 (P35372) | Homo sapiens | S268 | LKSVRMLSGSKEKDR | 0.385 | 0.0909 | - | - | - | - | 10908300 | - |
|
| SNCA (P37840) | Homo sapiens | S129 | NEAYEMPSEEGYQDY | 0.253 | 0.8853 | - | - | - | - | - | - |
|
| GRIA1 * (P42261 ) | Homo sapiens | S567 | FSPYEWHSEEFEEGR | 0.054 | 0.3182 | TP | certain | experimental | 73 | 8 | 72 | 55 | 74 | | 21135237 | - |
|
| CDKN1B (P46527) | Homo sapiens | S10 | NVRVSNGSPSLERMD | 0.644 | 0.7415 | TP | certain | experimental | 73 | 8 | 74 | | 20851109 | - |
|
| Rph3a (P47709) | Rattus norvegicus | S274 | QGLRRANSVQASRPA | - | 0.9711 | TP | certain | experimental | 73 | 8 | 55 | | 11466417 | - |
|
| VIM (P48616) | Bos taurus | S26 | GTASRPSSTRSYVTT | - | 0.7718 | - | - | - | - | - | - |
|
| VIM (P48616) | Bos taurus | S39 | TTSTRTYSLGSALRP | - | 0.4801 | - | - | - | - | - | - |
|
| VIM (P48616) | Bos taurus | S412 | EGEESRISLPLPNFS | - | 0.4256 | - | - | - | - | - | - |
|
| VIM (P48616) | Bos taurus | S66 | GVYATRSSAVRLRSG | - | 0.5254 | - | - | - | - | - | - |
|
| VIM (P48616) | Bos taurus | S72 | SSAVRLRSGVPGVRL | - | 0.4409 | - | - | - | - | - | - |
|
| RRAD (P55042) | Homo sapiens | S273 | AGTRRRESLGKKAKR | 0.915 | 0.494 | TP | certain | experimental | 73 | 321 | 9 | | 9677319 | - |
|
| PLN (P61012) | Canis lupus familiaris | S16 | RSAIRRASTIEMPQQ | - | 0.0888 | - | - | - | - | - | - |
|
| Cdk5r1 (P61810) | Rattus norvegicus | S91 | ENLKKSLSCANLSTF | - | 0.3529 | TP | certain | experimental | 8 | 73 | 205 | 55 | 72 | | 20097924 | - |
|
| Mecp2 (Q00566) | Rattus norvegicus | S421 | EKMPRAGSLESDGCP | - | 0.8313 | TP | certain | experimental | 148 | 73 | 8 | 214 | 205 | | 17046689 | - |
|
| Grin2b (Q00960) | Rattus norvegicus | S1303 | NKLRRQHSYDTFVDL | - | 0.5665 | TP | certain | experimental | 73 | 214 | 273 | 55 | | 8940188 | - |
|
| Cacna1c (Q01815) | Mus musculus | S1487 | DYLTRDWSILGPHHL | - | 0.0646 | TP | certain | experimental | 148 | 8 | 73 | | 20479240 | - |
|
| Cacna1c (Q01815) | Mus musculus | S1545 | VACKRLVSMNMPLNS | - | 0.2645 | TP | certain | experimental | 148 | 8 | 73 | | 20479240 | - |
|
| Cacna1b (Q02294) | Rattus norvegicus | S2126 | SEKQRFYSCDRFGSR | - | 0.725 | TP | certain | experimental | 8 | 55 | 148 | 73 | | 16982421 | - |
|
| Cacna1b (Q02294) | Rattus norvegicus | S784 | RLQNLRASCEALYSE | - | 0.4685 | TP | certain | experimental | 107 | 8 | | 15607937 | - |
|
| Cacna1b (Q02294) | Rattus norvegicus | S896 | ERARPRRSHSKEAPG | - | 0.9634 | - | - | experimental | 107 | 8 | | 15607937 | - |
|
| GFPT1 (Q06210) | Homo sapiens | S261 | CNLSRVDSTTCLFPV | 0.557 | 0.3149 | - | - | - | - | - | - |
|
| GRIN2B (Q13224) | Homo sapiens | S1303 | NKLRRQHSYDTFVDL | 0.872 | 0.5758 | TP | certain | experimental | 73 | 8 | | 27998718 | - |
|
| ESPL1 (Q14674) | Homo sapiens | S1501 | TDNWRKMSFEILRGS | 0.927 | 0.5176 | - | - | - | - | - | - |
|
| SMAD2 (Q15796) | Homo sapiens | S110 | SFSEQTRSLDGRLQV | 0.488 | 0.3494 | TP | certain | experimental | 73 | 8 | 273 | 72 | 55 | | 11027280 | - |
|
| SMAD2 * (Q15796 ) | Homo sapiens | S240 | SDQQLNQSMDTGSPA | 0.323 | 0.8235 | TP | certain | experimental | 73 | 8 | 273 | 72 | 55 | | 11027280 | - |
|
| SMAD2 * (Q15796 ) | Homo sapiens | S260 | TLSPVNHSLDLQPVT | 0.145 | 0.5456 | TP | certain | experimental | 73 | 8 | 273 | 72 | 55 | | 11027280 | - |
|
| Dlg1 (Q62696) | Rattus norvegicus | S232 | ITLERGNSGLGFSIA | - | 0.374 | TP | certain | experimental | 72 | 73 | 8 | 74 | | 15044483 | - |
|
| Dlg1 (Q62696) | Rattus norvegicus | S39 | SSIERVISIFQSNLF | - | 0.1275 | TP | certain | experimental | 72 | 73 | 8 | 74 | | 15044483 | - |
|
| Kcnd2 * (Q63881 ) | Rattus norvegicus | S438 | ARIRAAKSGSANAYM | - | 0.3494 | TP | certain | experimental | 273 | 107 | 8 | 73 | 72 | | 15071113 | - |
|
| Kcnd2 * (Q63881 ) | Rattus norvegicus | S459 | LLSNQLQSSEDEPAF | - | 0.4619 | TP | certain | experimental | 273 | 107 | 8 | 73 | 72 | | 15071113 | - |
|
| Tph2 (Q8CGV2) | Mus musculus | S19 | YWARRGLSLDSAVPE | - | 0.4256 | TP | certain | experimental | 73 | 214 | 9 | 55 | | 17727633 | - |
|
| RYR2 * (Q92736 ) | Homo sapiens | S2814 | ISQTSQVSVDAAHGY | 0.345 | 0.494 | TP | certain | experimental | 72 | 55 | 62 | 23 | 55 | 72 | 148 | | 20056922; 15016728 | - |
|
| RCHY1 * (Q96PM5 ) | Homo sapiens | S155 | CLEDIHTSRVVAHVL | 0.847 | 0.0104 | TP | certain | experimental | 72 | 73 | 74 | 66 | 214 | 9 | | 17568776 | - |
|
| CARD11 * (Q9BXL7 ) | Homo sapiens | S116 | KEPTRRFSTIVVEEG | 0.795 | 0.4541 | - | - | - | 73 | 8 | | 16809782 | - |
|
| Rims1 (Q9JIR4) | Rattus norvegicus | S241 | RLQERSRSQTPLSTA | - | 0.7369 | TP | certain | experimental | 73 | 8 | | 12871946 | - |
|
| Rims1 (Q9JIR4) | Rattus norvegicus | S287 | KQASRSRSEPPRERK | - | 0.9577 | TP | certain | experimental | 73 | 8 | | 12871946 | - |
|
| Pdc (Q9QW08) | Mus musculus | S54 | KEILRQMSSPQSRDD | - | 0.7881 | TP | certain | experimental | 55 | 73 | | 17569665 | - |
|
| Lrp4 (Q9QYP1) | Rattus norvegicus | S1887 | TTPERRGSLPDTGWK | - | 0.7843 | TP | certain | experimental | 74 | 214 | 73 | | 16819975 | - |
|
| Lrp4 (Q9QYP1) | Rattus norvegicus | S1900 | WKHERKLSSESQV__ | - | 0.5758 | TP | certain | experimental | 74 | 214 | 73 | | 16819975 | - |
|
| Gabbr1 (Q9WV18) | Mus musculus | S867 | ITRGEWQSEAQDTMK | - | 0.5098 | TP | certain | experimental | 3 | 23 | 214 | 9 | 73 | 55 | 72 | | 20643921 | - |
|
| PEA15 (Q9Z297) | Cricetulus griseus | S116 | KDIIRQPSEEEIIKL | - | 0.4017 | TP | certain | experimental | 55 | 73 | 8 | | 15916534 | - |
|
| FBXO43 (Q4G163) | Homo sapiens | T234 | FSQQKTSTIDDSKDD | 0.945 | 0.3807 | - | - | - | - | - | - |
|
| Brsk1 * (B2DD29 ) | Rattus norvegicus | T189 | VGDSLLETSCGSPHY | - | 0.1702 | - | - | - | 214 | 73 | 8 | 427 | 72 | | 18324781 | - |
|
| SLN * (O00631 ) | Homo sapiens | T5 | ___MGINTRELFLNF | 0.223 | 0.0033 | TP | certain | experimental | 73 | 9 | | 19631655 | - |
|
| ITGB1BP1 (O14713) | Homo sapiens | T38 | GGLSRSSTVASLDTD | 0.272 | 0.4831 | TP | certain | experimental | 72 | 9 | 148 | | 9813144 | - |
|
| Trpv1 (O35433) | Rattus norvegicus | T704 | WKLQRAITILDTEKS | - | 0.0568 | TP | certain | experimental | 72 | 9 | | 14630912 | - |
|
| ITGB1 (P05556) | Homo sapiens | T788 | PIYKSAVTTVVNPKY | 0.400 | 0.3019 | - | - | - | - | - | - |
|
| ITGB1 (P05556) | Homo sapiens | T789 | IYKSAVTTVVNPKYE | 0.670 | 0.2817 | - | - | - | - | - | - |
|
| Chrm4 (P08485) | Rattus norvegicus | T145 | TYPARRTTKMAGLMI | - | 0.0568 | TP | certain | experimental | 73 | 3 | 8 | 23 | 72 | | 20461055 | - |
|
| Camk2a (P11275) | Rattus norvegicus | T305 | KLKGAILTTMLATRN | - | 0.2884 | TP | certain | experimental | 73 | 62 | 107 | | 1324926 | - |
|
| Camk2a * (P11275 ) | Rattus norvegicus | T306 | LKGAILTTMLATRNF | - | 0.3117 | TP | certain | experimental | 73 | 62 | 107 | | 1324926 | - |
|
| Camk2a * (P11275 ) | Rattus norvegicus | T310 | ILTTMLATRNFSGGK | - | 0.3948 | TP | certain | experimental | 73 | 62 | 107 | | 1324926 | - |
|
| CALD1 (P12957) | Gallus gallus | T484 | QNGERELTTPKLKST | - | 0.8462 | TP | certain | experimental | 73 | 107 | | 2170388 | - |
|
| TNNT2 (P13789) | Bos taurus | T191 | GYIQKAQTERKSGKR | - | 0.6531 | TP | certain | experimental | 73 | 133 | | 7748902 | - |
|
| PLN (P26678) | Homo sapiens | T17 | SAIRRASTIEMPQQA | 0.522 | 0.1671 | TP | certain | experimental | 72 | 55 | | 20056922 | - |
|
| Gjc1 * (P28229 ) | Mus musculus | T337 | HLPADLETLQREIRM | - | 0.5992 | TP | certain | experimental | 73 | 214 | | 22127232 | - |
|
| CHAT (P28329-3) | Homo sapiens | T456 | VDNIRSATPEALAFV | 0.290 | 0.1969 | TP | certain | experimental | 214 | 73 | 62 | 273 | 148 | | 12486117 | - |
|
| PLN (P61012) | Canis lupus familiaris | T17 | SAIRRASTIEMPQQA | - | 0.1759 | - | - | - | - | - | - |
|
| GLO1 * (Q04760 ) | Homo sapiens | T107 | ELTHNWGTEDDETQS | 0.487 | 0.4766 | TP | certain | experimental | 8 | 55 | 72 | | 19199007 | - |
|
| Myh9 (Q62812) | Rattus norvegicus | T1940 | RRIVRKGTGDCSDEE | - | 0.5176 | TP | certain | experimental | 72 | 148 | 73 | 273 | | 10945986 | - |
|
| Sln (Q6SLE7) | Rattus norvegicus | T5 | ___MERSTQELFINF | - | 0.0083 | - | - | - | - | - | - |
|
| RCHY1 * (Q96PM5 ) | Homo sapiens | T154 | ICLEDIHTSRVVAHV | 0.837 | 0.0108 | TP | certain | experimental | 72 | 73 | 74 | 66 | 214 | 9 | | 17568776 | - |
|
| Sln (Q9CQD6) | Mus musculus | T5 | ___MERSTQELFINF | - | 0.0083 | TP | certain | experimental | 73 | 9 | | 19631655 | - |
|
| CAMK2A (Q9UQM7) | Homo sapiens | T305 | KLKGAILTTMLATRN | 0.754 | 0.2849 | - | - | - | - | - | - |
|
| CAMK2A (Q9UQM7) | Homo sapiens | T306 | LKGAILTTMLATRNF | 0.607 | 0.3117 | - | - | - | - | - | - |
|
| (Q9Z305) | Cavia porcellus | T1603 | RTALRIKTEGNLEQA | - | 0.3392 | TP | certain | experimental | 73 | 8 | 55 | | 19340532 | - |
|
| NCOR2 * (Q9Y618 ) | Homo sapiens | S2426 | ASGDRPPSVSSVHSE | 0.177 | 0.7916 | - | - | - | - | - | - |
|
| NCOR2 (Q9Y618) | Homo sapiens | S2453 | VWEDRPSSAGSTPFP | 0.734 | 0.7415 | - | - | - | - | - | - |
|
| Itpr2 (P29995) | Rattus norvegicus | S150 | EKNAMRVSLDAAGNE | - | 0.2645 | TP | certain | experimental | 148 | 74 | 73 | 321 | 62 | 5572 | | 23019322 | - |
|
| HDAC5 (Q9UQL6) | Homo sapiens | S259 | FPLRKTASEPNLKVR | 0.835 | 0.6712 | - | - | - | - | - | - |
|
| Mcu (Q3UMR5) | Mus musculus | S91 | VISVRLPSRRERCQF | - | 0.1914 | - | - | - | - | - | - |
|
| Mybpc3 (O70468) | Mus musculus | S302 | SLLKKRDSFRRDSKL | - | 0.384 | - | - | - | - | - | - |
|
| TH (P07101-4) | Homo sapiens | S35 | AIMVRGQSPRFIGRR | 0.086 | 0.4619 | - | - | - | - | - | - |
|
| TH (P07101-4) | Homo sapiens | S44 | RFIGRRQSLIEDARK | 0.066 | 0.5758 | - | - | - | - | - | - |
|
| Kif3a (P28741) | Mus musculus | S698 | KPETVIDSLLQ____ | - | 0.4831 | - | - | - | - | - | - |
|
| Slc6a3 (P23977) | Rattus norvegicus | S13 | CSVGPMSSVVAPAKE | - | 0.3426 | - | - | - | - | - | - |
|
| STMN1 (P16949) | Homo sapiens | S16 | KELEKRASGQAFELI | 0.337 | 0.4087 | - | - | - | - | - | - |
|
| Cpeb3 (Q7TN99) | Mus musculus | S298 | SPLKKPFSSNVIAPP | - | 0.5017 | - | - | - | - | - | - |
|
| KIF3A (Q9Y496) | Homo sapiens | S687 | KTGRRKRSAKPETVI | 0.388 | 0.662 | - | - | - | - | - | - |
|
| Cpeb3 (Q7TN99) | Mus musculus | S420 | GRRRGRSSLFPFEDA | - | 0.2752 | - | - | - | - | - | - |
|
| Ppp3ca (P63329) | Rattus norvegicus | S411 | GKMARVFSVLREESE | - | 0.2224 | - | - | - | - | - | - |
|
| TH (P07101-4) | Homo sapiens | S19 | KGFRRAVSELDAKQA | 0.455 | 0.4766 | - | - | - | - | - | - |
|
| DAGLA (Q9Y4D2) | Homo sapiens | S782 | APLATMESLSDTESL | 0.651 | 0.4725 | - | - | - | - | - | - |
|
| Abi1 (Q8CBW3) | Mus musculus | S88 | SQLRRMESSINHISQ | - | 0.4652 | - | - | - | - | - | - |
|
| Mcu (Q3UMR5) | Mus musculus | S56 | TAHQRPASWQSVGAA | - | 0.3529 | - | - | - | - | - | - |
|
| HDAC5 (Q9UQL6) | Homo sapiens | S498 | RPLSRTQSSPLPQSP | 0.940 | 0.9126 | - | - | - | - | - | - |
|
| NOX5 (Q96PH1-4) | Homo sapiens | S475 | WTNRLYESFKASDPL | 0.820 | 0.2817 | - | - | - | - | - | - |
|
| SLC6A4 (P31645) | Homo sapiens | S13 | LNSQKQLSACEDGED | 0.186 | 0.6043 | - | - | - | - | - | - |
|
| Syngap1 (Q9QUH6) | Rattus norvegicus | S1138 | PSITKQHSQTPSTLN | - | 0.892 | - | - | - | - | - | - |
|
| KCNQ2 (O43526-4) | Homo sapiens | S438 | QTVRRSPSADQSLED | 0.977 | 0.8198 | - | - | - | - | - | - |
|
| Neto2 (C6K2K4) | Rattus norvegicus | S409 | SLREKEISADLADLS | - | 0.2328 | - | - | - | - | - | - |
|
| Cpeb3 (Q7TN99) | Mus musculus | S299 | PLKKPFSSNVIAPPK | - | 0.4979 | - | - | - | - | - | - |
|
| CTNNB1 (P35222) | Homo sapiens | S552 | QDTQRRTSMGGTQQQ | 0.142 | 0.6661 | - | - | - | - | - | - |
|
| BECN1 (Q14457) | Homo sapiens | S90 | IPPARMMSTESANSF | 0.467 | 0.4582 | - | - | - | - | - | - |
|
| DLGAP1 (O14490-2) | Homo sapiens | S44 | CRRMRSGSYIKAMGD | 0.890 | 0.5176 | - | - | - | - | - | - |
|
| Wee2 (Q66JT0) | Mus musculus | S15 | QGLNKKLSFSFCEED | - | 0.6755 | - | - | - | - | - | - |
|
| ID1 (P41134) | Homo sapiens | S36 | GEVVRCLSEQSVAIS | 0.692 | 0.1702 | - | - | - | - | - | - |
|
| Fosb (P13346) | Mus musculus | S27 | SAESQYLSSVDSFGS | - | 0.662 | - | - | - | - | - | - |
|
| NR1H2 (P55055) | Homo sapiens | S426 | RMLMKLVSLRTLSSV | 0.159 | 0.1048 | - | - | - | - | - | - |
|
| DAGLA (Q9Y4D2) | Homo sapiens | S808 | RSIRGSPSLHAVLER | 0.387 | 0.5577 | - | - | - | - | - | - |
|
| Psmc5 (P62198) | Rattus norvegicus | S120 | RVALRNDSYTLHKIL | - | 0.3215 | - | - | - | - | - | - |
|
| Cpeb3 (Q7TN99) | Mus musculus | S291 | PSPLNPISPLKKPFS | - | 0.5098 | - | - | - | - | - | - |
|
| Cpeb3 (Q7TN99) | Mus musculus | S443 | QALSSGLSSPTRCQN | - | 0.662 | - | - | - | - | - | - |
|
| Cpeb3 (Q7TN99) | Mus musculus | S419 | YGRRRGRSSLFPFED | - | 0.2715 | - | - | - | - | - | - |
|
| TCAP (O15273) | Homo sapiens | S161 | RSLSRSMSQEAQRG_ | 0.844 | 0.5665 | - | - | - | - | - | - |
|
| Oprm1 (P33535) | Rattus norvegicus | T370 | STRVRQNTREHPSTA | - | 0.6991 | - | - | - | - | - | - |
|
| KCNJ11 (Q14654) | Homo sapiens | T180 | QAHRRAETLIFSKHA | 0.891 | 0.0888 | - | - | - | - | - | - |
|
| Fosb (P13346) | Mus musculus | T180 | RNRRRELTDRLQAET | - | 0.7289 | - | - | - | - | - | - |
|
| Kif3a (P28741) | Mus musculus | T694 | KRSAKPETVIDSLLQ | - | 0.5456 | - | - | - | - | - | - |
|
| Fosb (P13346) | Mus musculus | T149 | PRRPREETLTPEEEE | - | 0.9269 | - | - | - | - | - | - |
|
| Grm1 (P23385) | Rattus norvegicus | T871 | KLPCRSNTFLNIFRR | - | 0.2988 | - | - | - | - | - | - |
|
| SLC1A3 (P43003) | Homo sapiens | T26 | QQGVRKRTLLAKKKV | 0.297 | 0.2503 | - | - | - | - | - | - |
|
| Trpc6 (Q61143) | Mus musculus | T487 | RQLFRMKTSCFSWME | - | 0.0168 | - | - | - | - | - | - |
|
| SLC1A3 (P43003) | Homo sapiens | T37 | KKKVQNITKEDVKSY | 0.186 | 0.0704 | - | - | - | - | - | - |
|
| Dpysl2 (O08553) | Mus musculus | T555 | DNIPRRTTQRIVAPP | - | 0.7547 | - | - | - | - | - | - |
|
| Cpeb3 (Q7TN99) | Mus musculus | T446 | SSGLSSPTRCQNGER | - | 0.5901 | - | - | - | - | - | - |
|
| CTNNB1 (P35222) | Homo sapiens | T472 | ICALRHLTSRHQEAE | 0.915 | 0.3704 | - | - | - | - | - | - |
|
| CTNNB1 (P35222) | Homo sapiens | T332 | VNIMRTYTYEKLLWT | 0.009 | 0.0568 | - | - | - | - | - | - |
|
| KCNJ11 (Q14654) | Homo sapiens | T224 | MQVVRKTTSPEGEVV | 0.299 | 0.2951 | - | - | - | - | - | - |
|
| TNNT2 (P45379-2) | Homo sapiens | T203 | YIQKQAQTERKSGKR | - | 0.6375 | - | - | - | - | - | - | - |
| DLG1 (Q12959-8) | Homo sapiens | S116 | ITLERGNSGLGFSIA | 0.359 | 0.3529 | - | - | - | - | - | - | - |
| GABRG2 (P18507-2) | Homo sapiens | S382 | NPLLRMFSFKAPTID | 0.352 | 0.0909 | - | - | - | - | - | - | - |
| PDC (P20941-3) | Homo sapiens | S91 | KEILRQMSSPQSRNG | 0.215 | 0.805 | - | - | - | - | - | - | - |
| CLCN3 (P51790-5) | Homo sapiens | S51 | NGGSINSSTHLLDLL | 0.227 | 0.374 | - | - | - | - | - | - | - |
| PTPRA (P18433-6) | Homo sapiens | S180 | QAGSHSNSFRLSNGR | 0.044 | 0.5901 | - | - | - | - | - | - | - |
| BCL10 (O95999) | Homo sapiens | S138 | NNLSRSNSDESNFSE | 0.846 | 0.6482 | - | - | - | - | - | - | - |
| KRT18 (P05783) | Homo sapiens | S53 | ISVSRSTSFRGGMGS | 0.418 | 0.4476 | - | - | - | - | - | - | - |
| MAPT (P10636-2) | Homo sapiens | S267 | RVQSKIGSLDNITHV | 0.744 | 0.5211 | - | - | - | - | - | - | - |
| MAPT (P10636-6) | Homo sapiens | S204 | NVKSKIGSTENLKHQ | 0.668 | 0.8125 | - | - | - | - | - | - | - |
| MAPT (P10636-6) | Homo sapiens | S298 | RVQSKIGSLDNITHV | 0.868 | 0.4831 | - | - | - | - | - | - | - |
| MAPT (P10636-8) | Homo sapiens | S356 | RVQSKIGSLDNITHV | 0.096 | 0.4831 | - | - | - | - | - | - | - |
| CREB1 (P16220) | Homo sapiens | S142 | RKILNDLSSDAPGVP | - | 0.5707 | - | - | - | - | - | - | - |
| ATP2A2 (P16615-2) | Homo sapiens | S38 | KLKERWGSNELPAEE | 0.070 | 0.4149 | - | - | - | - | - | - | - |
| PTPRA (P18433) | Homo sapiens | S204 | TEDVEPQSVPLLARS | 0.557 | 0.712 | - | - | - | - | - | - | - |
| PDC (P20941-2) | Homo sapiens | S21 | ERVSRKMSIQEYELI | 0.978 | 0.4119 | - | - | - | - | - | - | - |
| DRD3 (P35462) | Homo sapiens | S229 | RILTRQNSQCNSVRP | 0.873 | 0.433 | - | - | - | - | - | - | - |
| GRIA1 (P42261) | Homo sapiens | S832 | LIEFCYKSRSESKRM | 0.458 | 0.1007 | - | - | - | - | - | - | - |
| DLG1 (Q12959) | Homo sapiens | S232 | ITLERGNSGLGFSIA | 0.126 | 0.3529 | - | - | - | - | - | - | - |
| IRF3 (Q14653) | Homo sapiens | S386 | ARVGGASSLENTVDL | 0.981 | 0.3149 | - | - | - | - | - | - | - |
| ABI1 (Q8IZP0) | Homo sapiens | S88 | SQLRRMESSINHISQ | 0.782 | 0.4685 | - | - | - | - | - | - | - |
| HOMER3 (Q9NSC5) | Homo sapiens | S120 | ARLAREKSQDGGELT | 0.386 | 0.6851 | - | - | - | - | - | - | - |
| HOMER3 (Q9NSC5) | Homo sapiens | S159 | EKLFRSQSADAPGPT | 0.688 | 0.8681 | - | - | - | - | - | - | - |
| HOMER3 (Q9NSC5) | Homo sapiens | S176 | ERLKKMLSEGSVGEV | 0.328 | 0.5342 | - | - | - | - | - | - | - |
| RYR2 (H7BY35) | Homo sapiens | S2792 | YNRTRRISQTSQVSV | - | 0.346 | - | - | - | - | - | - | - |
| TRPV1 (Q8NER1) | Homo sapiens | S502 | YFLQRRPSMKTLFVD | 0.412 | 0.0102 | - | - | - | - | - | - | - |
| SYN1 (P17600) | Homo sapiens | S605 | AGPTRQASQAGPVPR | 0.334 | 0.9905 | - | - | - | - | - | - | - |
| NEFL (P07196) | Homo sapiens | S58 | LSVRRSYSSSSGSLM | 0.185 | 0.3948 | - | - | - | - | - | - | - |
| NEFL (P07196) | Homo sapiens | S67 | SSGSLMPSLENLDLS | 0.173 | 0.3704 | - | - | - | - | - | - | - |
| CACNA1S (Q13698) | Homo sapiens | S1575 | PEICRTVSGDLAAEE | - | 0.4685 | - | - | - | - | - | - | - |
| (Q53FJ6) | Homo sapiens | S244 | KDRVKGKSDPYHATS | - | 0.384 | - | - | - | - | - | - | - |
| (Q53FJ6) | Homo sapiens | S330 | AGSTISNSHAQPFDF | - | 0.6991 | - | - | - | - | - | - | - |
| (Q53FJ6) | Homo sapiens | S364 | AIVDQRPSSRASSRA | - | 0.7951 | - | - | - | - | - | - | - |
| (Q53FJ6) | Homo sapiens | S365 | IVDQRPSSRASSRAS | - | 0.8085 | - | - | - | - | - | - | - |
| (Q53FJ6) | Homo sapiens | S369 | RPSSRASSRASSRPR | - | 0.9346 | - | - | - | - | - | - | - |
| (Q53FJ6) | Homo sapiens | S372 | SRASSRASSRPRPDD | - | 0.8765 | - | - | - | - | - | - | - |
| (Q53FJ6) | Homo sapiens | S373 | RASSRASSRPRPDDL | - | 0.8741 | - | - | - | - | - | - | - |
| (Q53FJ6) | Homo sapiens | S255 | HATSGALSPAKDCGS | - | 0.3249 | - | - | - | - | - | - | - |
| (Q53FJ6) | Homo sapiens | S296 | VTGDRNNSSCRNYNK | - | 0.4409 | - | - | - | - | - | - | - |
| (Q53FJ6) | Homo sapiens | S297 | TGDRNNSSCRNYNKQ | - | 0.5456 | - | - | - | - | - | - | - |
| (Q53FJ6) | Homo sapiens | S306 | RNYNKQASEQNWANY | - | 0.433 | - | - | - | - | - | - | - |
| (Q53FJ6) | Homo sapiens | S314 | EQNWANYSAEQNRMG | - | 0.6269 | - | - | - | - | - | - | - |
| (Q53FJ6) | Homo sapiens | S325 | NRMGQAGSTISNSHA | - | 0.7289 | - | - | - | - | - | - | - |
| (Q53FJ6) | Homo sapiens | S328 | GQAGSTISNSHAQPF | - | 0.6806 | - | - | - | - | - | - | - |
| MPZ (P25189) | Homo sapiens | S210 | HKPGKDASKRGRQTP | 0.909 | 0.5017 | - | - | - | - | - | - | - |
| MPZ (P25189) | Homo sapiens | S233 | SRSTKAVSEKKAKGL | 0.397 | 0.4864 | - | - | - | - | - | - | - |
| CAMK2A (Q7LDD5) | Homo sapiens | S314 | MLATRNFSGGKSGGN | - | 0.4979 | - | - | - | - | - | - | - |
| CALD1 (E7EX44) | Homo sapiens | S408 | CFTPKGSSLKIEERA | - | 0.4149 | - | - | - | - | - | - | - |
| CALD1 (E7EX44) | Homo sapiens | S441 | AIVSKIDSRLEQYTS | - | 0.5098 | - | - | - | - | - | - | - |
| CALD1 (Q05682) | Homo sapiens | S73 | VEVNAQNSVPDEEAK | 0.782 | 0.8765 | - | - | - | - | - | - | - |
| CALD1 (E7EX44) | Homo sapiens | S547 | RNLWEKQSVDKVTSP | - | 0.6712 | - | - | - | - | - | - | - |
| (A8K8W7) | Homo sapiens | S855 | EPMRRSVSEAALAQP | - | 0.7415 | - | - | - | - | - | - | - |
| CREB1 (P16220) | Homo sapiens | S133 | EILSRRPSYRKILND | - | 0.4979 | - | - | - | - | - | - | - |
| CACNA1C (Q13936) | Homo sapiens | S409 | GVLSGEFSKEREKAK | 0.737 | 0.2849 | - | - | - | - | - | - | - |
| CACNA1C (F5H522) | Homo sapiens | S1512 | DYLTRDWSILGPHHL | - | 0.0646 | - | - | - | - | - | - | - |
| CACNA1C (F5H522) | Homo sapiens | S1570 | VACKRLVSMNMPLNS | - | 0.2645 | - | - | - | - | - | - | - |
| GRIA1 (P42261) | Homo sapiens | S645 | LTVERMVSPIESAED | 0.029 | 0.1275 | - | - | - | - | - | - | - |
| GRIA1 (P42261) | Homo sapiens | S849 | FCLIPQQSINEAIRT | 0.238 | 0.1852 | - | - | - | - | - | - | - |
| GRIA4 (P48058) | Homo sapiens | S862 | IRNKARLSITGSVGE | 0.708 | 0.3872 | - | - | - | - | - | - | - |
| (A8K0K0) | Homo sapiens | S849 | FCLIPQQSINEAMRT | - | 0.2364 | - | - | - | - | - | - | - |
| GJC1 (P36383) | Homo sapiens | S326 | AQEQQYGSHEENLPA | 0.743 | 0.6948 | - | - | - | - | - | - | - |
| GJC1 (P36383) | Homo sapiens | S381 | SKAGSNKSTASSKSG | 0.680 | 0.7951 | - | - | - | - | - | - | - |
| GJC1 (P36383) | Homo sapiens | S384 | GSNKSTASSKSGDGK | 0.738 | 0.7982 | - | - | - | - | - | - | - |
| GJC1 (P36383) | Homo sapiens | S385 | SNKSTASSKSGDGKT | 0.824 | 0.7036 | - | - | - | - | - | - | - |
| GJC1 (P36383) | Homo sapiens | S387 | KSTASSKSGDGKTSV | 0.830 | 0.6427 | - | - | - | - | - | - | - |
| GJC1 (P36383) | Homo sapiens | S393 | KSGDGKTSVWI____ | 0.988 | 0.5139 | - | - | - | - | - | - | - |
| MAPT (A0A0G2JS76) | Homo sapiens | S751 | SNVSSTGSIDMVDSP | - | 0.5577 | - | - | - | - | - | - | - |
| NOS1 (C9J5P6) | Homo sapiens | S851 | SYKVRFNSVSSYSDS | - | 0.346 | - | - | - | - | - | - | - |
| VIM (P08670) | Homo sapiens | S39 | TTSTRTYSLGSALRP | 0.934 | 0.4652 | - | - | - | - | - | - | - |
| DLG4 (P78352) | Homo sapiens | S73 | ITLERGNSGLGFSIA | 0.221 | 0.3631 | - | - | - | - | - | - | - |
| OPRM1 (P35372) | Homo sapiens | S263 | LMILRLKSVRMLSGS | 0.189 | 0.0136 | - | - | - | - | - | - | - |
| PTK2 (Q05397) | Homo sapiens | S843 | DVRLSRGSIDREDGS | 0.494 | 0.6851 | - | - | - | - | - | - | - |
| RPH3A (Q9Y2J0) | Homo sapiens | S272 | AGLRRANSVQASRPA | 0.914 | 0.9649 | - | - | - | - | - | - | - |
| VIM (P08670) | Homo sapiens | S26 | GTASRPSSSRSYVTT | 0.396 | 0.7547 | - | - | - | - | - | - | - |
| VIM (P08670) | Homo sapiens | S412 | EGEESRISLPLPNFS | 0.035 | 0.422 | - | - | - | - | - | - | - |
| VIM (P08670) | Homo sapiens | S66 | GVYATRSSAVRLRSS | 0.166 | 0.5098 | - | - | - | - | - | - | - |
| VIM (P08670) | Homo sapiens | S72 | SSAVRLRSSVPGVRL | 0.756 | 0.4256 | - | - | - | - | - | - | - |
| GABRB1 (P18505) | Homo sapiens | S409 | IQYRKPLSSREAYGR | 0.088 | 0.2364 | - | - | - | - | - | - | - |
| GABRB1 (P18505) | Homo sapiens | S434 | GRIRRRASQLKVKIP | 0.418 | 0.4476 | - | - | - | - | - | - | - |
| PLN (P26678) | Homo sapiens | S16 | RSAIRRASTIEMPQQ | 0.748 | 0.0851 | - | - | - | - | - | - | - |
| CDK5R1 (Q8TAM4) | Homo sapiens | S91 | ENLKKSLSCANLSTF | - | 0.3631 | - | - | - | - | - | - | - |
| IL6ST (P40189) | Homo sapiens | S782 | QVFSRSESTQPLLDS | 0.885 | 0.6531 | - | - | - | - | - | - | - |
| MECP2 (E9LUH4) | Homo sapiens | S423 | EKMPRGGSLESDGCP | - | 0.8343 | - | - | - | - | - | - | - |
| CACNA1B (F6RH32) | Homo sapiens | S2130 | SEKQRFYSCDRFGGR | - | 0.7718 | - | - | - | - | - | - | - |
| CACNA1B (F6RH32) | Homo sapiens | S784 | RLQNLRASCEALYSE | - | 0.4476 | - | - | - | - | - | - | - |
| CACNA1B (F6RH32) | Homo sapiens | S892 | ERPRPHRSHSKEAAG | - | 0.9801 | - | - | - | - | - | - | - |
| KCND2 (A4D0V9) | Homo sapiens | S438 | ARIRAAKSGSANAYM | - | 0.346 | - | - | - | - | - | - | - |
| KCND2 (A4D0V9) | Homo sapiens | S459 | LLSNQLQSSEDEQAF | - | 0.4369 | - | - | - | - | - | - | - |
| TPH2 (Q8IWU9) | Homo sapiens | S19 | YWARRGFSLDSAVPE | 0.500 | 0.4119 | - | - | - | - | - | - | - |
| RIMS1 (Q86UR5) | Homo sapiens | S218 | RLQERSRSQTPLSTA | 0.999 | 0.7718 | - | - | - | - | - | - | - |
| RIMS1 (Q86UR5) | Homo sapiens | S265 | QASSRSRSEPPRERK | 0.867 | 0.9488 | - | - | - | - | - | - | - |
| LRP4 (O75096) | Homo sapiens | S1887 | ATPERRGSLPDTGWK | 0.500 | 0.7415 | - | - | - | - | - | - | - |
| LRP4 (O75096) | Homo sapiens | S1900 | WKHERKLSSESQV__ | 0.000 | 0.5533 | - | - | - | - | - | - | - |
| (A0A1U9X7R0) | Homo sapiens | S868 | ITRGEWQSEAQDTMK | - | 0.5139 | - | - | - | - | - | - | - |
| PEA15 (Q15121) | Homo sapiens | S116 | KDIIRQPSEEEIIKL | 0.538 | 0.4017 | - | - | - | - | - | - | - |
| (A9CB80) | Homo sapiens | S1064 | SCPIKEDSFLQRYSS | - | 0.5055 | - | - | - | - | - | - | - |
| (A9CB80) | Homo sapiens | S1081 | TGALTEDSIDDTFLP | - | 0.7163 | - | - | - | - | - | - | - |
| (A9CB80) | Homo sapiens | S1120 | QPLNPAPSRDPHYQD | - | 0.8623 | - | - | - | - | - | - | - |
| GFAP (K7EMP8) | Homo sapiens | S13 | ITSAARRSYVSSGEM | - | 0.4292 | - | - | - | - | - | - | - |
| GFAP (K7EMP8) | Homo sapiens | S17 | ARRSYVSSGEMMVGG | - | 0.4441 | - | - | - | - | - | - | - |
| GFAP (K7EMP8) | Homo sapiens | S38 | LGPGTRLSLARMPPP | - | 0.6851 | - | - | - | - | - | - | - |
| GFAP (K7EMP8) | Homo sapiens | S393 | NLQIRETSLDTKSVS | - | 0.5176 | - | - | - | - | - | - | - |
| (Q5U0M2) | Homo sapiens | S282 | NSLQRVPSYDSFDSE | - | 0.3667 | - | - | - | - | - | - | - |
| PDC (P20941) | Homo sapiens | S73 | ERVSRKMSIQEYELI | 0.490 | 0.4507 | - | - | - | - | - | - | - |
| SYNGAP1 (A0A1U9X8L0) | Homo sapiens | S1073 | PPLQRGKSQQLTVSA | - | 0.9168 | - | - | - | - | - | - | - |
| MCU (Q8NE86) | Homo sapiens | S92 | VISVRLPSRRERCQF | 0.936 | 0.1852 | - | - | - | - | - | - | - |
| MYBPC3 (A8MXZ9) | Homo sapiens | S304 | SLLKKSSSFRTPRDS | - | 0.3631 | - | - | - | - | - | - | - |
| KIF3A (E9PES4) | Homo sapiens | S723 | KPETVIDSLLQ____ | - | 0.4831 | - | - | - | - | - | - | - |
| SLC6A3 (Q01959) | Homo sapiens | S13 | CSVGLMSSVVAPAKE | 0.500 | 0.2609 | - | - | - | - | - | - | - |
| CPEB3 (Q5QP71) | Homo sapiens | S297 | SPLKKPFSSNVIAPP | - | 0.4979 | - | - | - | - | - | - | - |
| CPEB3 (Q5QP71) | Homo sapiens | S388 | MALNSRSSLFPFEDA | - | 0.2002 | - | - | - | - | - | - | - |
| PPP3CA (A0A0S2Z4C6) | Homo sapiens | S411 | GKMARVFSVLREESE | - | 0.2258 | - | - | - | - | - | - | - |
| MCU (Q8NE86) | Homo sapiens | S57 | TVHQRIASWQNLGAV | 0.336 | 0.1583 | - | - | - | - | - | - | - |
| SYNGAP1 (A0A1U9X8L0) | Homo sapiens | S1138 | PSITKQHSQTPSTLN | - | 0.892 | - | - | - | - | - | - | - |
| CPEB3 (Q5QP71) | Homo sapiens | S298 | PLKKPFSSNVIAPPK | - | 0.4901 | - | - | - | - | - | - | - |
| FOSB (A0A024R0P6) | Homo sapiens | S27 | SAESQYLSSVDSFGS | - | 0.6712 | - | - | - | - | - | - | - |
| (A0A140VJS3) | Homo sapiens | S120 | RVALRNDSYTLHKIL | - | 0.3215 | - | - | - | - | - | - | - |
| CPEB3 (Q5QP71) | Homo sapiens | S290 | PSPLNPISPLKKPFS | - | 0.5017 | - | - | - | - | - | - | - |
| CPEB3 (Q5QP71) | Homo sapiens | S411 | QALSSGLSSPTRCQN | - | 0.662 | - | - | - | - | - | - | - |
| CPEB3 (Q5QP71) | Homo sapiens | S387 | IMALNSRSSLFPFED | - | 0.1942 | - | - | - | - | - | - | - |
| ATP2A2 (P16615) | Homo sapiens | S38 | KLKERWGSNELPAEE | 0.070 | 0.4149 | - | - | - | - | - | - | - |
| CEBPB (P17676) | Homo sapiens | S325 | EQLSRELSTLRNLFK | 0.002 | 0.5296 | - | - | - | - | - | - | - |
| CFL2 (Q9Y281) | Homo sapiens | S24 | DMKVRKSSTQEEIKK | 0.207 | 0.3774 | - | - | - | - | - | - | - |
| ADCY3 (O60266) | Homo sapiens | S1076 | NVASRMESTGVMGNI | 0.605 | 0.3321 | - | - | - | - | - | - | - |
| QDPR (P09417) | Homo sapiens | S223 | ITGKNRPSSGSLIQV | 0.149 | 0.4087 | - | - | - | - | - | - | - |
| GFAP (P14136) | Homo sapiens | S38 | LGPGTRLSLARMPPP | 0.368 | 0.6851 | - | - | - | - | - | - | - |
| GFAP (P14136) | Homo sapiens | S13 | ITSAARRSYVSSGEM | 0.795 | 0.4292 | - | - | - | - | - | - | - |
| GFAP (P14136) | Homo sapiens | S17 | ARRSYVSSGEMMVGG | 0.894 | 0.4441 | - | - | - | - | - | - | - |
| KCND2 (Q9NZV8) | Homo sapiens | S438 | ARIRAAKSGSANAYM | 0.059 | 0.346 | - | - | - | - | - | - | - |
| TH (P07101) | Homo sapiens | S44 | PGPSLTGSPWPGTAA | 0.066 | 0.8198 | - | - | - | - | - | - | - |
| SYN1 (P17600) | Homo sapiens | S568 | PQATRQTSVSGPAPP | 0.924 | 0.9898 | - | - | - | - | - | - | - |
| ACACA (Q13085) | Homo sapiens | S25 | RFIIGSVSEDNSEDE | 0.000 | 0.4901 | - | - | - | - | - | - | - |
| RYR1 (P21817) | Homo sapiens | S2843 | KKKTRKISQSAQTYD | 0.703 | 0.6851 | - | - | - | - | - | - | - |
| GRIN2B (Q13224) | Homo sapiens | S383 | VGKWKDKSLQMKYYV | 0.724 | 0.1007 | - | - | - | - | - | - | - |
| PPP1R9B (D3DTX6) | Homo sapiens | S116 | ALLKLGTSVSERVSR | - | 0.5176 | - | - | - | - | - | - | - |
| PPP1R9B (D3DTX6) | Homo sapiens | S100 | LSLPRASSLNENVDH | - | 0.6136 | - | - | - | - | - | - | - |
| MYLK (Q15746-7) | Homo sapiens | S838 | RAIGRLSSMAMISGL | 0.291 | 0.3286 | - | - | - | - | - | - | - |
| ITGB1BP1 (O14713-2) | Homo sapiens | T38 | GGLSRSSTVASLDTD | 0.272 | 0.4901 | - | - | - | - | - | - | - |
| CHAT (P28329) | Homo sapiens | T574 | VDNIRSATPEALAFV | 0.465 | 0.1969 | - | - | - | - | - | - | - |
| BRSK1 (Q8TDC3) | Homo sapiens | T189 | VGDSLLETSCGSPHY | 0.685 | 0.1702 | - | - | - | - | - | - | - |
| TRPV1 (Q8NER1) | Homo sapiens | T705 | WKLQRAITILDTEKS | 0.539 | 0.0555 | - | - | - | - | - | - | - |
| CHRM4 (Q96RG8) | Homo sapiens | T146 | TYPARRTTKMAGLMI | - | 0.0555 | - | - | - | - | - | - | - |
| CAMK2A (Q7LDD5) | Homo sapiens | T305 | KLKGAILTTMLATRN | - | 0.2849 | - | - | - | - | - | - | - |
| CAMK2A (Q7LDD5) | Homo sapiens | T306 | LKGAILTTMLATRNF | - | 0.3117 | - | - | - | - | - | - | - |
| CAMK2A (Q7LDD5) | Homo sapiens | T310 | ILTTMLATRNFSGGK | - | 0.391 | - | - | - | - | - | - | - |
| MYF4 (Q9HCA4) | Homo sapiens | T87 | VDRRRAATLREKRRL | - | 0.4441 | - | - | - | - | - | - | - |
| SLN (A0A158RFT9) | Homo sapiens | T5 | ___MGINTRELFLNF | - | 0.0033 | - | - | - | - | - | - | - |
| nlgn1 (D2X2H5) | Homo sapiens | T759 | CSPQRTTTNDLTHAQ | - | 0.8565 | - | - | - | - | - | - | - |
| CAMK2A (Q7LDD5) | Homo sapiens | T286 | SCMHRQETVDCLKKF | - | 0.2752 | - | - | - | - | - | - | - |
| OPRM1 (P35372) | Homo sapiens | T372 | STRIRQNTRDHPSTA | 0.461 | 0.6712 | - | - | - | - | - | - | - |
| FOSB (A0A024R0P6) | Homo sapiens | T180 | RNRRRELTDRLQAET | - | 0.7505 | - | - | - | - | - | - | - |
| KIF3A (E9PES4) | Homo sapiens | T719 | KRSAKPETVIDSLLQ | - | 0.5456 | - | - | - | - | - | - | - |
| FOSB (A0A024R0P6) | Homo sapiens | T149 | PRRPREETLTPEEEE | - | 0.9308 | - | - | - | - | - | - | - |
| GRM1 (Q13255) | Homo sapiens | T871 | KLPCRSNTFLNIFRR | - | 0.2364 | - | - | - | - | - | - | - |
| TRPC6 (Q9Y210) | Homo sapiens | T488 | KQLFRMKTSCFSWME | 0.457 | 0.0168 | - | - | - | - | - | - | - |
| (Q53ET2) | Homo sapiens | T555 | DNIPRRTTQRIVAPP | - | 0.7547 | - | - | - | - | - | - | - |
| CPEB3 (Q5QP71) | Homo sapiens | T414 | SSGLSSPTRCQNGER | - | 0.5854 | - | - | - | - | - | - | - |
| VPS72 (Q15906) | Homo sapiens | T86 | RRKRRVVTKAYKEPL | 0.905 | 0.6136 | - | - | - | - | - | - | - |
| VPS72 (Q15906) | Homo sapiens | T137 | RKSMRQSTAEHTRQT | 0.990 | 0.7718 | - | - | - | - | - | - | - |
| VPS72 (Q15906) | Homo sapiens | T168 | PHCERPLTQEELLRE | 0.927 | 0.6427 | - | - | - | - | - | - | - |
| (Q53H78) | Homo sapiens | T287 | SMMYRQETVECLKKF | - | 0.2645 | - | - | - | - | - | - | - |
Help Instructions
- Hovering on the "*" in "Substrate" column will display the text if there is a note by the annotator for the given site.
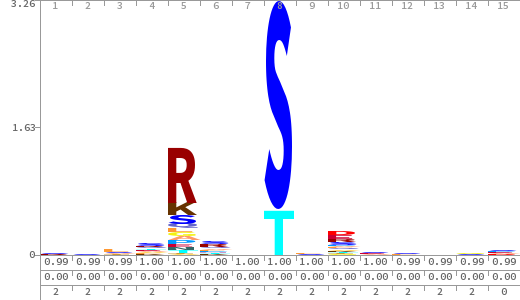
- The column having "Sequence" motif represents phosphosite and its neighbouring 7 residues (upstream and downstream).
- The score in "Conservation" column varies from 0 to 1; Higher the score more conserved is the site.
- The score in "Disorder" column varies from 0 to 1; Score higher than 0.4 suggests disorder.
- In the "Curator Assessment" column, TP refers to True Positive; FP refers to False Positive; and U refers to unknown if the annoatator cannot decide the status based on available information.
- Hovering on the number in the "Evidence Logic" column will show the name of method used to characterize/infer the phosphosite (for more details plase follow the link)